Selection of specific protein binders for pre-defined targets from an optimized library of artificial helicoidal repeat proteins (alphaRep)
- PMID: 24014183
- PMCID: PMC3754942
- DOI: 10.1371/journal.pone.0071512
Selection of specific protein binders for pre-defined targets from an optimized library of artificial helicoidal repeat proteins (alphaRep)
Abstract
We previously designed a new family of artificial proteins named αRep based on a subgroup of thermostable helicoidal HEAT-like repeats. We have now assembled a large optimized αRep library. In this library, the side chains at each variable position are not fully randomized but instead encoded by a distribution of codons based on the natural frequency of side chains of the natural repeats family. The library construction is based on a polymerization of micro-genes and therefore results in a distribution of proteins with a variable number of repeats. We improved the library construction process using a "filtration" procedure to retain only fully coding modules that were recombined to recreate sequence diversity. The final library named Lib2.1 contains 1.7×10(9) independent clones. Here, we used phage display to select, from the previously described library or from the new library, new specific αRep proteins binding to four different non-related predefined protein targets. Specific binders were selected in each case. The results show that binders with various sizes are selected including relatively long sequences, with up to 7 repeats. ITC-measured affinities vary with Kd values ranging from micromolar to nanomolar ranges. The formation of complexes is associated with a significant thermal stabilization of the bound target protein. The crystal structures of two complexes between αRep and their cognate targets were solved and show that the new interfaces are established by the variable surfaces of the repeated modules, as well by the variable N-cap residues. These results suggest that αRep library is a new and versatile source of tight and specific binding proteins with favorable biophysical properties.
Conflict of interest statement
Figures
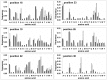
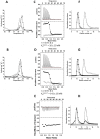
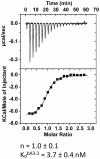

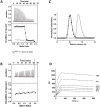
 ): elution profile of the GFP alone (6.75 nmol). (D) Affinity determination of selected bGFP-A using SPR. Different concentrations of bGFP-A (71,3; 118; 142,6; 237,6; 713; 1426 nM) were applied to flow cell with immobilized biotinylated EGFP for 120 s followed by washing buffer flow. The sensorgrams were corrected for non-specific binding by subtraction of a channel without EGFP bound (grey curve). The fits of kon and koff rates are indicated by black dashed line. Kd values were computed using koff = 1.7×10−4 s−1 for all concentrations and kon = 4.3, 4, 2.2, 2.6, 2, 2×104M−1 s−1 for the increasing concentrations respectively.
): elution profile of the GFP alone (6.75 nmol). (D) Affinity determination of selected bGFP-A using SPR. Different concentrations of bGFP-A (71,3; 118; 142,6; 237,6; 713; 1426 nM) were applied to flow cell with immobilized biotinylated EGFP for 120 s followed by washing buffer flow. The sensorgrams were corrected for non-specific binding by subtraction of a channel without EGFP bound (grey curve). The fits of kon and koff rates are indicated by black dashed line. Kd values were computed using koff = 1.7×10−4 s−1 for all concentrations and kon = 4.3, 4, 2.2, 2.6, 2, 2×104M−1 s−1 for the increasing concentrations respectively.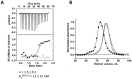


References
-
- Urvoas A, Valerio-Lepiniec M, Minard P (2012) Artificial proteins from combinatorial approaches. Trends Biotechnol 30: 512–520. - PubMed
-
- Hey T, Fiedler E, Rudolph R, Fiedler M (2005) Artificial, non-antibody binding proteins for pharmaceutical and industrial applications. Trends Biotechnol 23: 514–522. - PubMed
-
- Skerra A (2007) Alternative non-antibody scaffolds for molecular recognition. Curr Opin Biotechnol 18: 295–304. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

