Large Conformational Changes of Insertion 3 in Human Glycyl-tRNA Synthetase (hGlyRS) during Catalysis
- PMID: 26797133
- PMCID: PMC4786711
- DOI: 10.1074/jbc.M115.679126
Large Conformational Changes of Insertion 3 in Human Glycyl-tRNA Synthetase (hGlyRS) during Catalysis
Abstract
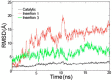
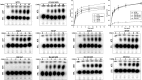
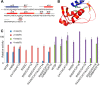
Glycyl-tRNA synthetase (GlyRS) is the enzyme that covalently links glycine to cognate tRNA for translation. It is of great research interest because of its nonconserved quaternary structures, unique species-specific aminoacylation properties, and noncanonical functions in neurological diseases, but none of these is fully understood. We report two crystal structures of human GlyRS variants, in the free form and in complex with tRNA(Gly) respectively, and reveal new aspects of the glycylation mechanism. We discover that insertion 3 differs considerably in conformation in catalysis and that it acts like a "switch" and fully opens to allow tRNA to bind in a cross-subunit fashion. The flexibility of the protein is supported by molecular dynamics simulation, as well as enzymatic activity assays. The biophysical and biochemical studies suggest that human GlyRS may utilize its flexibility for both the traditional function (regulate tRNA binding) and alternative functions (roles in diseases).
Keywords: Charcot-Marie-Tooth disease (CMT); aminoacyl tRNA synthetase; conformational change; crystal structure; enzyme mechanism.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures




References
-
- Eriani G., Delarue M., Poch O., Gangloff J., and Moras D. (1990) Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature 347, 203–206 - PubMed
-
- Schimmel P. (1987) Aminoacyl tRNA synthetases: general scheme of structure-function relationships in the polypeptides and recognition of transfer RNAs. Annu. Rev. Biochem. 56, 125–158 - PubMed
-
- Cusack S. (1993) Sequence, structure and evolutionary relationships between class 2 aminoacyl-tRNA synthetases: an update. Biochimie 75, 1077–1081 - PubMed
-
- Perona J. J., and Gruic-Sovulj I. (2014) Synthetic and editing mechanisms of aminoacyl-tRNA synthetases. Top. Curr. Chem. 344, 1–41 - PubMed
-
- Srinivasan G., James C. M., and Krzycki J. A. (2002) Pyrrolysine encoded by UAG in Archaea: charging of a UAG-decoding specialized tRNA. Science 296, 1459–1462 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

