Structure, function, and mechanism of the core circadian clock in cyanobacteria
- PMID: 29440392
- PMCID: PMC5892564
- DOI: 10.1074/jbc.TM117.001433
Structure, function, and mechanism of the core circadian clock in cyanobacteria
Abstract
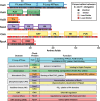
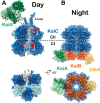
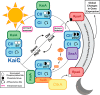
Circadian rhythms enable cells and organisms to coordinate their physiology with the cyclic environmental changes that come as a result of Earth's light/dark cycles. Cyanobacteria make use of a post-translational oscillator to maintain circadian rhythms, and this elegant system has become an important model for circadian timekeeping mechanisms. Composed of three proteins, the KaiABC system undergoes an oscillatory biochemical cycle that provides timing cues to achieve a 24-h molecular clock. Together with the input/output proteins SasA, CikA, and RpaA, these six gene products account for the timekeeping, entrainment, and output signaling functions in cyanobacterial circadian rhythms. This Minireview summarizes the current structural, functional and mechanistic insights into the cyanobacterial circadian clock.
Keywords: ATPases associated with diverse cellular activities (AAA); bacterial protein kinase; bacterial signal transduction; circadian rhythm; crystallography; nuclear magnetic resonance (NMR).
© 2018 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures

References
-
- Mitsui A., Kumazawa S., Takahashi A., Ikemoto H., Cao S., and Arai T. (1986) Strategy by which nitrogen-fixing unicellular cyanobacteria grow photoautotrophically. Nature 323, 720–722 10.1038/323720a0 - DOI
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions

