Negative Epistasis and Evolvability in TEM-1 β-Lactamase--The Thin Line between an Enzyme's Conformational Freedom and Disorder
- PMID: 26004540
- PMCID: PMC4718737
- DOI: 10.1016/j.jmb.2015.05.011
Negative Epistasis and Evolvability in TEM-1 β-Lactamase--The Thin Line between an Enzyme's Conformational Freedom and Disorder
Abstract
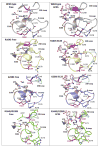
Epistasis is a key factor in evolution since it determines which combinations of mutations provide adaptive solutions and which mutational pathways toward these solutions are accessible by natural selection. There is growing evidence for the pervasiveness of sign epistasis--a complete reversion of mutational effects, particularly in protein evolution--yet its molecular basis remains poorly understood. We describe the structural basis of sign epistasis between G238S and R164S, two adaptive mutations in TEM-1 β-lactamase--an enzyme that endows antibiotics resistance. Separated by 10 Å, these mutations initiate two separate trajectories toward increased hydrolysis rates and resistance toward second and third-generation cephalosporins antibiotics. Both mutations allow the enzyme's active site to adopt alternative conformations and accommodate the new antibiotics. By solving the corresponding set of crystal structures, we found that R164S causes local disorder whereas G238S induces discrete conformations. When combined, the mutations in 238 and 164 induce local disorder whereby nonproductive conformations that perturb the enzyme's catalytic preorganization dominate. Specifically, Asn170 that coordinates the deacylating water molecule is misaligned, in both the free form and the inhibitor-bound double mutant. This local disorder is not restored by stabilizing global suppressor mutations and thus leads to an evolutionary cul-de-sac. Conformational dynamism therefore underlines the reshaping potential of protein's structures and functions but also limits protein evolvability because of the fragility of the interactions networks that maintain protein structures.
Keywords: conformational diversity; interactions network; protein disorder; protein evolution; protein folds.
Copyright © 2015 Elsevier Ltd. All rights reserved.
Figures


References
-
- Breen MS, Kemena C, Vlasov PK, Notredame C, Kondrashov FA. Epistasis as the primary factor in molecular evolution. Nature. 490:535–8. - PubMed
-
- de Visser JA, Krug J. Empirical fitness landscapes and the predictability of evolution. Nat Rev Genet. 2014;15:480–90. - PubMed
-
- Weinreich DM, Delaney NF, Depristo MA, Hartl DL. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science. 2006;312:111–4. - PubMed
-
- Borman AM, Paulous S, Clavel F. Resistance of human immunodeficiency virus type 1 to protease inhibitors: selection of resistance mutations in the presence and absence of the drug. J Gen Virol. 1996;77(Pt 3):419–26. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

