Probing the metastable state of influenza hemagglutinin
- PMID: 29127198
- PMCID: PMC5766969
- DOI: 10.1074/jbc.M117.815043
Probing the metastable state of influenza hemagglutinin
Abstract
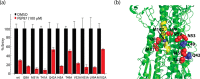
Viral entry into host cells is mediated by membrane proteins in a metastable state that transition to a more stable state upon a stimulus. For example, in the influenza envelope protein hemagglutinin (HA), the low pH in the endosome triggers a transition from the metastable prefusion conformation to the stable fusion conformation. To identify probes that interfere with HA function, here we screened a library of H7 HA peptides for inhibition of H7 HA-mediated entry. We discovered a peptide, PEP87 (WSYNAELLVAMENQHTI), that inhibited H7 and H5 HA-mediated entry. PEP87 corresponds to a highly conserved helical region of the HA2 subunit of HA that self-interacts in the neutral pH conformation. Mutagenesis experiments indicated that PEP87 binds to its native region in the HA trimer. We also found that PEP87 is unstructured in isolation but tends to form a helix as evidenced by CD and NMR studies. Fluorescence, chemical cross-linking, and saturation transfer difference NMR data suggested that PEP87 binds to the neutral pH conformation of HA and disrupts the HA structure without affecting its oligomerization state. Together, this work provides support for a model in which PEP87 disrupts HA function by displacing native interactions of the neutral pH conformation. Moreover, our observations indicate that the HA prefusion structure (and perhaps the metastable states of other viral entry proteins) is more dynamic with transient motions being larger than generally appreciated. These findings also suggest that the ensemble of prefusion structures presents many potential sites for targeting in therapeutic interventions.
Keywords: glycoprotein structure; hemagglutinin; influenza virus; nuclear magnetic resonance (NMR); peptide conformation; virus.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

