Crystal structure of subunits D and F in complex gives insight into energy transmission of the eukaryotic V-ATPase from Saccharomyces cerevisiae
- PMID: 25505269
- PMCID: PMC4318993
- DOI: 10.1074/jbc.M114.622688
Crystal structure of subunits D and F in complex gives insight into energy transmission of the eukaryotic V-ATPase from Saccharomyces cerevisiae
Abstract
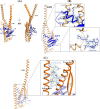
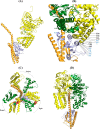
Eukaryotic V1VO-ATPases hydrolyze ATP in the V1 domain coupled to ion pumping in VO. A unique mode of regulation of V-ATPases is the reversible disassembly of V1 and VO, which reduces ATPase activity and causes silencing of ion conduction. The subunits D and F are proposed to be key in these enzymatic processes. Here, we describe the structures of two conformations of the subunit DF assembly of Saccharomyces cerevisiae (ScDF) V-ATPase at 3.1 Å resolution. Subunit D (ScD) consists of a long pair of α-helices connected by a short helix ((79)IGYQVQE(85)) as well as a β-hairpin region, which is flanked by two flexible loops. The long pair of helices is composed of the N-terminal α-helix and the C-terminal helix, showing structural alterations in the two ScDF structures. The entire subunit F (ScF) consists of an N-terminal domain of four β-strands (β1-β4) connected by four α-helices (α1-α4). α1 and β2 are connected via the loop (26)GQITPETQEK(35), which is unique in eukaryotic V-ATPases. Adjacent to the N-terminal domain is a flexible loop, followed by a C-terminal α-helix (α5). A perpendicular and extended conformation of helix α5 was observed in the two crystal structures and in solution x-ray scattering experiments, respectively. Fitted into the nucleotide-bound A3B3 structure of the related A-ATP synthase from Enterococcus hirae, the arrangements of the ScDF molecules reflect their central function in ATPase-coupled ion conduction. Furthermore, the flexibility of the terminal helices of both subunits as well as the loop (26)GQITPETQEK(35) provides information about the regulatory step of reversible V1VO disassembly.
Keywords: ATP Synthase; Bioenergetics; Proton Pump; Saccharomyces cerevisiae; Vacuolar ATPase.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures





References
-
- Kluge C., Lahr J., Hanitzsch M., Bolte S., Golldack D., Dietz K. J. (2003) New insight into the structure and regulation of the plant vacuolar H+-ATPase. J. Bioenerg. Biomembr. 35, 377–388 - PubMed
-
- Saroussi S., Nelson N. (2009) Vacuolar H+-ATPase–an enzyme for all seasons. Pflügers Arch. 457, 581–587 - PubMed
-
- Beyenbach K. W., Wieczorek H. (2006) The V-type H+ ATPase: molecular structure and function, physiological roles and regulation. J. Exp. Biol. 209, 577–589 - PubMed
-
- Nishi T., Forgac M. (2002) The vacuolar (H+)-ATPases–nature's most versatile proton pumps. Nat. Rev. Mol. Cell Biol. 3, 94–103 - PubMed
-
- Marshansky V., Rubinstein J. L., Grüber G. (2014) Eukaryotic V-ATPase: novel structural findings and functional insights. Biochim. Biophys. Acta 1837, 857–879 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

