Clathrin adaptors. AP2 controls clathrin polymerization with a membrane-activated switch
- PMID: 25061211
- PMCID: PMC4333214
- DOI: 10.1126/science.1254836
Clathrin adaptors. AP2 controls clathrin polymerization with a membrane-activated switch
Abstract
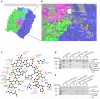
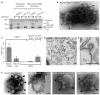
Clathrin-mediated endocytosis (CME) is vital for the internalization of most cell-surface proteins. In CME, plasma membrane-binding clathrin adaptors recruit and polymerize clathrin to form clathrin-coated pits into which cargo is sorted. Assembly polypeptide 2 (AP2) is the most abundant adaptor and is pivotal to CME. Here, we determined a structure of AP2 that includes the clathrin-binding β2 hinge and developed an AP2-dependent budding assay. Our findings suggest that an autoinhibitory mechanism prevents clathrin recruitment by cytosolic AP2. A large-scale conformational change driven by the plasma membrane phosphoinositide phosphatidylinositol 4,5-bisphosphate and cargo relieves this autoinhibition, triggering clathrin recruitment and hence clathrin-coated bud formation. This molecular switching mechanism can couple AP2's membrane recruitment to its key functions of cargo and clathrin binding.
Copyright © 2014, American Association for the Advancement of Science.
Figures


Comment in
-
Endocytosis: Unlocking AP2 activity.Nat Rev Mol Cell Biol. 2014 Sep;15(9):560-1. doi: 10.1038/nrm3863. Nat Rev Mol Cell Biol. 2014. PMID: 25145845 No abstract available.
References
-
- Dannhauser PN, Ungewickell EJ. Reconstitution of clathrin-coated bud and vesicle formation with minimal components. Nat. Cell Biol. 2012;14:634–639. - PubMed
-
- Collins BM, McCoy AJ, Kent HM, Evans PR, Owen DJ. Molecular architecture and functional model of the endocytic AP2 complex. Cell. 2002;109:523–535. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

