Structural Insights into Binding of the Antifungal Drug Fluconazole to Saccharomyces cerevisiae Lanosterol 14α-Demethylase
- PMID: 26055382
- PMCID: PMC4505223
- DOI: 10.1128/AAC.00925-15
Structural Insights into Binding of the Antifungal Drug Fluconazole to Saccharomyces cerevisiae Lanosterol 14α-Demethylase
Abstract
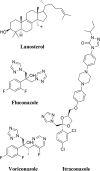
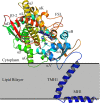
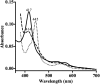
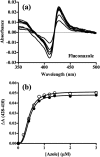
Infections by fungal pathogens such as Candida albicans and Aspergillus fumigatus and their resistance to triazole drugs are major concerns. Fungal lanosterol 14α-demethylase belongs to the CYP51 class in the cytochrome P450 superfamily of enzymes. This monospanning bitopic membrane protein is involved in ergosterol biosynthesis and is the primary target of azole antifungal drugs, including fluconazole. The lack of high-resolution structural information for this drug target from fungal pathogens has been a limiting factor for the design of modified triazole drugs that will overcome resistance. Here we report the X-ray structure of full-length Saccharomyces cerevisiae lanosterol 14α-demethylase in complex with fluconazole at a resolution of 2.05 Å. This structure shows the key interactions involved in fluconazole binding and provides insight into resistance mechanisms by revealing a water-mediated hydrogen bonding network between the drug and tyrosine 140, a residue frequently found mutated to histidine or phenylalanine in resistant clinical isolates.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures

References
-
- Horn DL, Fishman JA, Steinbach WJ, Anaissie EJ, Marr KA, Olyaei AJ, Pfaller MA, Weiss MA, Webster KM, Neofytos D. 2007. Presentation of the PATH Alliance registry for prospective data collection and analysis of the epidemiology, therapy, and outcomes of invasive fungal infections. Diagn Microbiol Infect Dis 59:407–414. doi:10.1016/j.diagmicrobio.2007.06.008. - DOI - PubMed
-
- US Department of Health and Human Services, Centers for Disease Control and Prevention. 2013. Antibiotic resistance threats in the United States. US Department of Health and Human Services, Centers for Disease Control and Prevention. http://www.cdc.gov/drugresistance/threat-report-2013/pdf/ar-threats-2013....
-
- Alexander BD, Johnson MD, Pfeiffer CD, Jimenez-Ortigosa C, Catania J, Booker R, Castanheira M, Messer SA, Perlin DS, Pfaller MA. 2013. Increasing echinocandin resistance in Candida glabrata: clinical failure correlates with presence of FKS mutations and elevated minimum inhibitory concentrations. Clin Infect Dis 56:1724–1732. doi:10.1093/cid/cit136. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

