Crystal structures capture three states in the catalytic cycle of a pyridoxal phosphate (PLP) synthase
- PMID: 25568319
- PMCID: PMC4342443
- DOI: 10.1074/jbc.M114.626382
Crystal structures capture three states in the catalytic cycle of a pyridoxal phosphate (PLP) synthase
Abstract
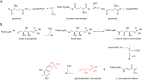
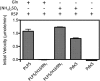
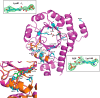
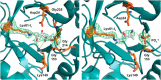
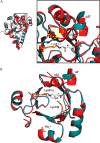
PLP synthase (PLPS) is a remarkable single-enzyme biosynthetic pathway that produces pyridoxal 5'-phosphate (PLP) from glutamine, ribose 5-phosphate, and glyceraldehyde 3-phosphate. The intact enzyme includes 12 synthase and 12 glutaminase subunits. PLP synthesis occurs in the synthase active site by a complicated mechanism involving at least two covalent intermediates at a catalytic lysine. The first intermediate forms with ribose 5-phosphate. The glutaminase subunit is a glutamine amidotransferase that hydrolyzes glutamine and channels ammonia to the synthase active site. Ammonia attack on the first covalent intermediate forms the second intermediate. Glyceraldehyde 3-phosphate reacts with the second intermediate to form PLP. To investigate the mechanism of the synthase subunit, crystal structures were obtained for three intermediate states of the Geobacillus stearothermophilus intact PLPS or its synthase subunit. The structures capture the synthase active site at three distinct steps in its complicated catalytic cycle, provide insights into the elusive mechanism, and illustrate the coordinated motions within the synthase subunit that separate the catalytic states. In the intact PLPS with a Michaelis-like intermediate in the glutaminase active site, the first covalent intermediate of the synthase is fully sequestered within the enzyme by the ordering of a generally disordered 20-residue C-terminal tail. Following addition of ammonia, the synthase active site opens and admits the Lys-149 side chain, which participates in formation of the second intermediate and PLP. Roles are identified for conserved Asp-24 in the formation of the first intermediate and for conserved Arg-147 in the conversion of the first to the second intermediate.
Keywords: Enzyme Structure; Glutaminase; Pyridoxal Phosphate; Vitamin; X-ray Crystallography.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures

References
-
- Eliot A. C., Kirsch J. F. (2004) Pyridoxal phosphate enzymes: mechanistic, structural, and evolutionary considerations. Annu. Rev. Biochem. 73, 383–415 - PubMed
-
- Christen P., Mehta P. K. (2001) From cofactor to enzymes. The molecular evolution of pyridoxal-5′-phosphate-dependent enzymes. Chem. Rec. 1, 436–447 - PubMed
-
- Jain S. K., Lim G. (2001) Pyridoxine and pyridoxamine inhibits superoxide radicals and prevents lipid peroxidation, protein glycosylation, and (Na+ + K+)-ATPase activity reduction in high glucose-treated human erythrocytes. Free Radic. Biol. Med. 30, 232–237 - PubMed
-
- Bilski P., Li M. Y., Ehrenshaft M., Daub M. E., Chignell C. F. (2000) Vitamin B6 (pyridoxine) and its derivatives are efficient singlet oxygen quenchers and potential fungal antioxidants. Photochem. Photobiol. 71, 129–134 - PubMed
-
- Ehrenshaft M., Jenns A. E., Chung K. R., Daub M. E. (1998) SOR1, a gene required for photosensitizer and singlet oxygen resistance in Cercospora fungi, is highly conserved in divergent organisms. Mol. Cell 1, 603–609 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

