UbiX is a flavin prenyltransferase required for bacterial ubiquinone biosynthesis
- PMID: 26083743
- PMCID: PMC4988493
- DOI: 10.1038/nature14559
UbiX is a flavin prenyltransferase required for bacterial ubiquinone biosynthesis
Abstract
Ubiquinone (also known as coenzyme Q) is a ubiquitous lipid-soluble redox cofactor that is an essential component of electron transfer chains. Eleven genes have been implicated in bacterial ubiquinone biosynthesis, including ubiX and ubiD, which are responsible for decarboxylation of the 3-octaprenyl-4-hydroxybenzoate precursor. Despite structural and biochemical characterization of UbiX as a flavin mononucleotide (FMN)-binding protein, no decarboxylase activity has been detected. Here we report that UbiX produces a novel flavin-derived cofactor required for the decarboxylase activity of UbiD. UbiX acts as a flavin prenyltransferase, linking a dimethylallyl moiety to the flavin N5 and C6 atoms. This adds a fourth non-aromatic ring to the flavin isoalloxazine group. In contrast to other prenyltransferases, UbiX is metal-independent and requires dimethylallyl-monophosphate as substrate. Kinetic crystallography reveals that the prenyltransferase mechanism of UbiX resembles that of the terpene synthases. The active site environment is dominated by π systems, which assist phosphate-C1' bond breakage following FMN reduction, leading to formation of the N5-C1' bond. UbiX then acts as a chaperone for adduct reorientation, via transient carbocation species, leading ultimately to formation of the dimethylallyl C3'-C6 bond. Our findings establish the mechanism for formation of a new flavin-derived cofactor, extending both flavin and terpenoid biochemical repertoires.
Conflict of interest statement
The authors declare no competing financial interest. Readers are welcome to comment on the online version of the paper.
Figures
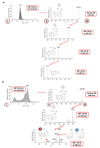
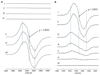

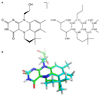


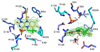
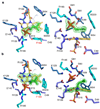


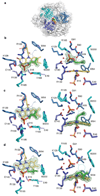
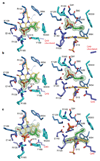

References
-
- Lenaz G, Genova ML. Mobility and function of coenzyme Q (ubiquinone) in the mitochondiral respiratory chain. Biochim Biophys Acta. 2009;1787:563–573. - PubMed
-
- Aussel L, Pierrel F, Loiseau L, Lombard M, Fontecave M, Barras F. Biosynthesis and physiology of coenzyme Q in bacteria. Biochim Biophys Acta. 2014;1837:1004–1011. - PubMed
-
- Payne KAP, et al. A new cofactor supports reversible decarboxylation of α, β-unsaturated acids reminiscent of 1,3-dipolar cycloaddition chemistry. Nature. in press. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
- BB/E013007/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/H021523/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/K017802/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/M017702/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

