Design, Synthesis, and Evaluation of Polyamine Deacetylase Inhibitors, and High-Resolution Crystal Structures of Their Complexes with Acetylpolyamine Amidohydrolase
- PMID: 26200446
- PMCID: PMC4526345
- DOI: 10.1021/acs.biochem.5b00536
Design, Synthesis, and Evaluation of Polyamine Deacetylase Inhibitors, and High-Resolution Crystal Structures of Their Complexes with Acetylpolyamine Amidohydrolase
Abstract
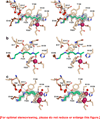
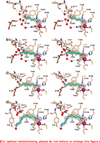
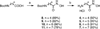
Polyamines are essential aliphatic polycations that bind to nucleic acids and accordingly are involved in a variety of cellular processes. Polyamine function can be regulated by acetylation and deacetylation, just as histone function can be regulated by lysine acetylation and deacetylation. Acetylpolyamine amidohydrolase (APAH) from Mycoplana ramosa is a zinc-dependent polyamine deacetylase that shares approximately 20% amino acid sequence identity with human histone deacetylases. We now report the X-ray crystal structures of APAH-inhibitor complexes in a new and superior crystal form that diffracts to very high resolution (1.1-1.4 Å). Inhibitors include previously synthesized analogues of N(8)-acetylspermidine bearing trifluoromethylketone, thiol, and hydroxamate zinc-binding groups [Decroos, C., Bowman, C. M., and Christianson, D. W. (2013) Bioorg. Med. Chem. 21, 4530], and newly synthesized hydroxamate analogues of shorter, monoacetylated diamines, the most potent of which is the hydroxamate analogue of N-acetylcadaverine (IC50 = 68 nM). The high-resolution crystal structures of APAH-inhibitor complexes provide key inferences about the inhibition and catalytic mechanism of zinc-dependent deacetylases. For example, the trifluoromethylketone analogue of N(8)-acetylspermidine binds as a tetrahedral gem-diol that mimics the tetrahedral intermediate and its flanking transition states in catalysis. Surprisingly, this compound is also a potent inhibitor of human histone deacetylase 8 with an IC50 of 260 nM. Crystal structures of APAH-inhibitor complexes are determined at the highest resolution of any currently existing zinc deacetylase structure and thus represent the most accurate reference points for understanding structure-mechanism and structure-inhibition relationships in this critically important enzyme family.
Conflict of interest statement
The authors declare no competing conflicts of interest.
Figures


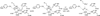
References
-
- Walsh CT, Garneau-Tsodikova S, Gatto GJ., Jr Protein posttranslational modifications: the chemistry of proteome diversifications. Angew. Chem., Int. Ed. 2005;44:7342–7372. - PubMed
-
- Workman JL, Kingston RE. Alteration of nucleosome as a mechanism of transcriptional regulation. Annu. Rev. Biochem. 1998;67:545–579. - PubMed
-
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets complexes and co-regulates major cellular functions. Science. 2009;325:834–840. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information
Research Materials

