i-bodies, Human Single Domain Antibodies That Antagonize Chemokine Receptor CXCR4
- PMID: 27036939
- PMCID: PMC4933451
- DOI: 10.1074/jbc.M116.721050
i-bodies, Human Single Domain Antibodies That Antagonize Chemokine Receptor CXCR4
Abstract
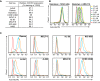
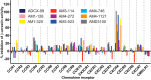
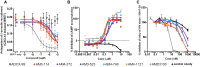
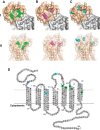
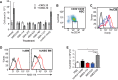
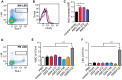
CXCR4 is a G protein-coupled receptor with excellent potential as a therapeutic target for a range of clinical conditions, including stem cell mobilization, cancer prognosis and treatment, fibrosis therapy, and HIV infection. We report here the development of a fully human single-domain antibody-like scaffold termed an "i-body," the engineering of which produces an i-body library possessing a long complementarity determining region binding loop, and the isolation and characterization of a panel of i-bodies with activity against human CXCR4. The CXCR4-specific i-bodies show antagonistic activity in a range of in vitro and in vivo assays, including inhibition of HIV infection, cell migration, and leukocyte recruitment but, importantly, not the mobilization of hematopoietic stem cells. Epitope mapping of the three CXCR4 i-bodies AM3-114, AM4-272, and AM3-523 revealed binding deep in the binding pocket of the receptor.
Keywords: CXC chemokine receptor type 4 (CXCR-4); G protein-coupled receptor (GPCR); antibody engineering; hematopoietic stem cells; protein structure; single-domain antibody (sdAb, nanobody).
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures
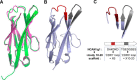

References
-
- Jost C., and Plückthun A. (2014) Engineered proteins with desired specificity: DARPins, other alternative scaffolds and bispecific IgGs. Curr. Opin. Struct. Biol. 27, 102–112 - PubMed
-
- Weidle U. H., Auer J., Brinkmann U., Georges G., and Tiefenthaler G. (2013) The emerging role of new protein scaffold-based agents for treatment of cancer. Cancer Genomics Proteomics 10, 155–168 - PubMed
-
- Wurch T., Pierré A., and Depil S. (2012) Novel protein scaffolds as emerging therapeutic proteins: from discovery to clinical proof-of-concept. Trends Biotechnol. 30, 575–582 - PubMed
-
- Rasmussen S. G., Choi H. J., Fung J. J., Pardon E., Casarosa P., Chae P. S., Devree B. T., Rosenbaum D. M., Thian F. S., Kobilka T. S., Schnapp A., Konetzki I., Sunahara R. K., Gellman S. H., Pautsch A., et al. (2011) Structure of a nanobody-stabilized active state of the β(2) adrenoceptor. Nature 469, 175–180 - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

