Directed evolution of the tryptophan synthase β-subunit for stand-alone function recapitulates allosteric activation
- PMID: 26553994
- PMCID: PMC4664345
- DOI: 10.1073/pnas.1516401112
Directed evolution of the tryptophan synthase β-subunit for stand-alone function recapitulates allosteric activation
Abstract
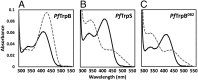
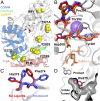
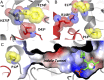
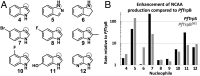
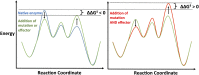
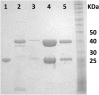
Enzymes in heteromeric, allosterically regulated complexes catalyze a rich array of chemical reactions. Separating the subunits of such complexes, however, often severely attenuates their catalytic activities, because they can no longer be activated by their protein partners. We used directed evolution to explore allosteric regulation as a source of latent catalytic potential using the β-subunit of tryptophan synthase from Pyrococcus furiosus (PfTrpB). As part of its native αββα complex, TrpB efficiently produces tryptophan and tryptophan analogs; activity drops considerably when it is used as a stand-alone catalyst without the α-subunit. Kinetic, spectroscopic, and X-ray crystallographic data show that this lost activity can be recovered by mutations that reproduce the effects of complexation with the α-subunit. The engineered PfTrpB is a powerful platform for production of Trp analogs and for further directed evolution to expand substrate and reaction scope.
Keywords: PLP; allostery; noncanonical amino acid; protein engineering.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



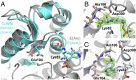
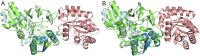
References
-
- Du L, Lou L. PKS and NRPS release mechanisms. Nat Prod Rep. 2010;27(2):255–278. - PubMed
-
- Hyde CC, Ahmed SA, Padlan EA, Miles EW, Davies DR. Three-dimensional structure of the tryptophan synthase alpha 2 beta 2 multienzyme complex from Salmonella typhimurium. J Biol Chem. 1988;263(33):17857–17871. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

