Structural basis for m7G recognition and 2'-O-methyl discrimination in capped RNAs by the innate immune receptor RIG-I
- PMID: 26733676
- PMCID: PMC4725518
- DOI: 10.1073/pnas.1515152113
Structural basis for m7G recognition and 2'-O-methyl discrimination in capped RNAs by the innate immune receptor RIG-I
Abstract
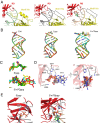
RNAs with 5'-triphosphate (ppp) are detected in the cytoplasm principally by the innate immune receptor Retinoic Acid Inducible Gene-I (RIG-I), whose activation triggers a Type I IFN response. It is thought that self RNAs like mRNAs are not recognized by RIG-I because 5'ppp is capped by the addition of a 7-methyl guanosine (m7G) (Cap-0) and a 2'-O-methyl (2'-OMe) group to the 5'-end nucleotide ribose (Cap-1). Here we provide structural and mechanistic basis for exact roles of capping and 2'-O-methylation in evading RIG-I recognition. Surprisingly, Cap-0 and 5'ppp double-stranded (ds) RNAs bind to RIG-I with nearly identical Kd values and activate RIG-I's ATPase and cellular signaling response to similar extents. On the other hand, Cap-0 and 5'ppp single-stranded RNAs did not bind RIG-I and are signaling inactive. Three crystal structures of RIG-I complexes with dsRNAs bearing 5'OH, 5'ppp, and Cap-0 show that RIG-I can accommodate the m7G cap in a cavity created through conformational changes in the helicase-motif IVa without perturbing the ppp interactions. In contrast, Cap-1 modifications abrogate RIG-I signaling through a mechanism involving the H830 residue, which we show is crucial for discriminating between Cap-0 and Cap-1 RNAs. Furthermore, m7G capping works synergistically with 2'-O-methylation to weaken RNA affinity by 200-fold and lower ATPase activity. Interestingly, a single H830A mutation restores both high-affinity binding and signaling activity with 2'-O-methylated dsRNAs. Our work provides new structural insights into the mechanisms of host and viral immune evasion from RIG-I, explaining the complexity of cap structures over evolution.
Keywords: RIG-I; capped RNA; crystal structure; innate immunity; self versus nonself.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Yoneyama M, et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol. 2005;175(5):2851–2858. - PubMed
-
- Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5(7):730–737. - PubMed
-
- Kowalinski E, et al. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147(2):423–435. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

