How repertoire data are changing antibody science
- PMID: 32409582
- PMCID: PMC7380193
- DOI: 10.1074/jbc.REV120.010181
How repertoire data are changing antibody science
Abstract
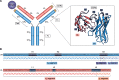
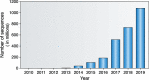
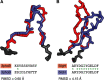
Antibodies are vital proteins of the immune system that recognize potentially harmful molecules and initiate their removal. Mammals can efficiently create vast numbers of antibodies with different sequences capable of binding to any antigen with high affinity and specificity. Because they can be developed to bind to many disease agents, antibodies can be used as therapeutics. In an organism, after antigen exposure, antibodies specific to that antigen are enriched through clonal selection, expansion, and somatic hypermutation. The antibodies present in an organism therefore report on its immune status, describe its innate ability to deal with harmful substances, and reveal how it has previously responded. Next-generation sequencing technologies are being increasingly used to query the antibody, or B-cell receptor (BCR), sequence repertoire, and the amount of BCR data in public repositories is growing. The Observed Antibody Space database, for example, currently contains over a billion sequences from 68 different studies. Repertoires are available that represent both the naive state (i.e. antigen-inexperienced) and that after immunization. This wealth of data has created opportunities to learn more about our immune system. In this review, we discuss the many ways in which BCR repertoire data have been or could be exploited. We highlight its utility for providing insights into how the naive immune repertoire is generated and how it responds to antigens. We also consider how structural information can be used to enhance these data and may lead to more accurate depictions of the sequence space and to applications in the discovery of new therapeutics.
Keywords: B-cell receptor (BCR); adaptive immunity; antibody; bioinformatics; immunology; next-generation sequencing; observed antibody space database; protein sequence; protein structure; structural annotation.
© 2020 Marks and Deane.
Conflict of interest statement
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
Figures

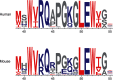
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

