Structural and functional consequences of a disease mutation in the telomere protein TPP1
- PMID: 27807141
- PMCID: PMC5135350
- DOI: 10.1073/pnas.1605685113
Structural and functional consequences of a disease mutation in the telomere protein TPP1
Abstract
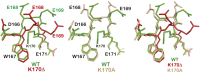
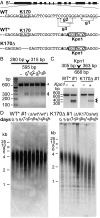
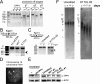
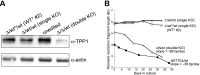
Telomerase replicates chromosome ends to facilitate continued cell division. Mutations that compromise telomerase function result in stem cell failure diseases, such as dyskeratosis congenita (DC). One such mutation (K170Δ), residing in the telomerase-recruitment factor TPP1, provides an excellent opportunity to structurally, biochemically, and genetically dissect the mechanism of such diseases. We show through site-directed mutagenesis and X-ray crystallography that this TPP1 disease mutation deforms the conformation of two critical amino acids of the TEL [TPP1's glutamate (E) and leucine-rich (L)] patch, the surface of TPP1 that binds telomerase. Using CRISPR-Cas9 technology, we demonstrate that introduction of this mutation in a heterozygous manner is sufficient to shorten telomeres in human cells. Our findings rule out dominant-negative effects of the mutation. Instead, these findings implicate reduced TEL patch dosage in causing telomere shortening. Our studies provide mechanistic insight into telomerase-deficiency diseases and encourage the development of gene therapies to counter such diseases.
Keywords: TEL patch; TPP1; dyskeratosis congenita; telomerase; telomere.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
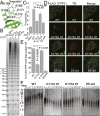
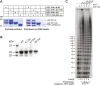
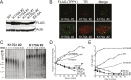
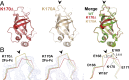


References
-
- Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43(2 Pt 1):405–413. - PubMed
-
- Meyerson M, et al. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell. 1997;90(4):785–795. - PubMed
-
- Lingner J, et al. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276(5312):561–567. - PubMed
-
- Savage SA. Human telomeres and telomere biology disorders. Prog Mol Biol Transl Sci. 2014;125:41–66. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

