The Structure of the Antibiotic Deactivating, N-hydroxylating Rifampicin Monooxygenase
- PMID: 27557658
- PMCID: PMC5076826
- DOI: 10.1074/jbc.M116.745315
The Structure of the Antibiotic Deactivating, N-hydroxylating Rifampicin Monooxygenase
Abstract
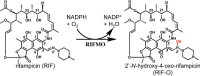
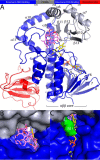
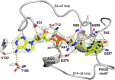
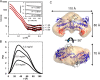
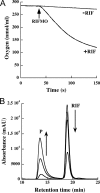
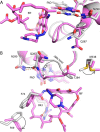
Rifampicin monooxygenase (RIFMO) catalyzes the N-hydroxylation of the natural product antibiotic rifampicin (RIF) to 2'-N-hydroxy-4-oxo-rifampicin, a metabolite with much lower antimicrobial activity. RIFMO shares moderate sequence similarity with well characterized flavoprotein monooxygenases, but the protein has not been isolated and characterized at the molecular level. Herein, we report crystal structures of RIFMO from Nocardia farcinica, the determination of the oligomeric state in solution with small angle x-ray scattering, and the spectrophotometric characterization of substrate binding. The structure identifies RIFMO as a class A flavoprotein monooxygenase and is similar in fold and quaternary structure to MtmOIV and OxyS, which are enzymes in the mithramycin and oxytetracycline biosynthetic pathways, respectively. RIFMO is distinguished from other class A flavoprotein monooxygenases by its unique middle domain, which is involved in binding RIF. Small angle x-ray scattering analysis shows that RIFMO dimerizes via the FAD-binding domain to form a bell-shaped homodimer in solution with a maximal dimension of 110 Å. RIF binding was monitored using absorbance at 525 nm to determine a dissociation constant of 13 μm Steady-state oxygen consumption assays show that NADPH efficiently reduces the FAD only when RIF is present, implying that RIF binds before NADPH in the catalytic scheme. The 1.8 Å resolution structure of RIFMO complexed with RIF represents the precatalytic conformation that occurs before formation of the ternary E-RIF-NADPH complex. The RIF naphthoquinone blocks access to the FAD N5 atom, implying that large conformational changes are required for NADPH to reduce the FAD. A model for these conformational changes is proposed.
Keywords: enzyme kinetics; enzyme structure; flavoprotein; small-angle X-ray scattering (SAXS); x-ray crystallography.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


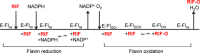

References
-
- Aristoff P. A., Garcia G. A., Kirchhoff P. D., and Hollis Showalter H. D. (2010) Rifamycins: obstacles and opportunities. Tuberculosis 90, 94–118 - PubMed
-
- Goldstein B. P. (2014) Resistance to rifampicin: a review. J. Antibiot. 67, 625–630 - PubMed
-
- Severinov K., Soushko M., Goldfarb A., and Nikiforov V. (1993) Rifampicin region revisited: new rifampicin-resistant and streptolydigin-resistant mutants in the β subunit of Escherichia coli RNA polymerase. J. Biol. Chem. 268, 14820–14825 - PubMed
-
- Telenti A., Imboden P., Marchesi F., Matter L., Schopfer K., Bodmer T., Lowrie D., Colston M. J., and Cole S. (1993) Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet 341, 647–650 - PubMed
-
- Tupin A., Gualtieri M., Roquet-Banères F., Morichaud Z., Brodolin K., and Leonetti J.-P.(2010) Resistance to rifampicin: at the crossroads between ecological, genomic and medical concerns. Int. J. Antimicrob. Agents 35, 519–523 - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

