An evolutionarily distinct family of polysaccharide lyases removes rhamnose capping of complex arabinogalactan proteins
- PMID: 28637865
- PMCID: PMC5555188
- DOI: 10.1074/jbc.M117.794578
An evolutionarily distinct family of polysaccharide lyases removes rhamnose capping of complex arabinogalactan proteins
Abstract
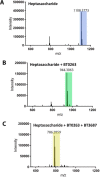
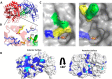
The human gut microbiota utilizes complex carbohydrates as major nutrients. The requirement for efficient glycan degrading systems exerts a major selection pressure on this microbial community. Thus, we propose that this microbial ecosystem represents a substantial resource for discovering novel carbohydrate active enzymes. To test this hypothesis we screened the potential enzymatic functions of hypothetical proteins encoded by genes of Bacteroides thetaiotaomicron that were up-regulated by arabinogalactan proteins or AGPs. Although AGPs are ubiquitous in plants, there is a paucity of information on their detailed structure, the function of these glycans in planta, and the mechanisms by which they are depolymerized in microbial ecosystems. Here we have discovered a new polysaccharide lyase family that is specific for the l-rhamnose-α1,4-d-glucuronic acid linkage that caps the side chains of complex AGPs. The reaction product generated by the lyase, Δ4,5-unsaturated uronic acid, is removed from AGP by a glycoside hydrolase located in family GH105, producing the final product 4-deoxy-β-l-threo-hex-4-enepyranosyl-uronic acid. The crystal structure of a member of the novel lyase family revealed a catalytic domain that displays an (α/α)6 barrel-fold. In the center of the barrel is a deep pocket, which, based on mutagenesis data and amino acid conservation, comprises the active site of the lyase. A tyrosine is the proposed catalytic base in the β-elimination reaction. This study illustrates how highly complex glycans can be used as a scaffold to discover new enzyme families within microbial ecosystems where carbohydrate metabolism is a major evolutionary driver.
Keywords: X-ray crystallography; carbohydrate processing; glycobiology; glycoside hydrolase; microbiome.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures



References
-
- Cuskin F., Lowe E. C., Temple M. J., Zhu Y., Cameron E. A., Pudlo N. A., Porter N. T., Urs K., Thompson A. J., Cartmell A., Rogowski A., Hamilton B. S., Chen R., Tolbert T. J., Piens K., et al. (2015) Human gut Bacteroidetes can utilize yeast mannan through a selfish mechanism. Nature 517, 165–169 - PMC - PubMed
-
- Larsbrink J., Rogers T. E., Hemsworth G. R., McKee L. S., Tauzin A. S., Spadiut O., Klinter S., Pudlo N. A., Urs K., Koropatkin N. M., Creagh A. L., Haynes C. A., Kelly A. G., Cederholm S. N., Davies G. J., Martens E. C., and Brumer H. (2014) A discrete genetic locus confers xyloglucan metabolism in select human gut Bacteroidetes. Nature 506, 498–502 - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

