The inactive C-terminal cassette of the dual-cassette RNA helicase BRR2 both stimulates and inhibits the activity of the N-terminal helicase unit
- PMID: 31914407
- PMCID: PMC7029114
- DOI: 10.1074/jbc.RA119.010964
The inactive C-terminal cassette of the dual-cassette RNA helicase BRR2 both stimulates and inhibits the activity of the N-terminal helicase unit
Abstract
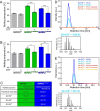
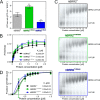
The RNA helicase bad response to refrigeration 2 homolog (BRR2) is required for the activation of the spliceosome before the first catalytic step of RNA splicing. BRR2 represents a distinct subgroup of Ski2-like nucleic acid helicases whose members comprise tandem helicase cassettes. Only the N-terminal cassette of BRR2 is an active ATPase and can unwind substrate RNAs. The C-terminal cassette represents a pseudoenzyme that can stimulate RNA-related activities of the N-terminal cassette. However, the molecular mechanisms by which the C-terminal cassette modulates the activities of the N-terminal unit remain elusive. Here, we show that N- and C-terminal cassettes adopt vastly different relative orientations in a crystal structure of BRR2 in complex with an activating domain of the spliceosomal Prp8 protein at 2.4 Å resolution compared with the crystal structure of BRR2 alone. Likewise, inspection of BRR2 structures within spliceosomal complexes revealed that the cassettes occupy different relative positions and engage in different intercassette contacts during different splicing stages. Engineered disulfide bridges that locked the cassettes in two different relative orientations had opposite effects on the RNA-unwinding activity of the N-terminal cassette, with one configuration enhancing and the other configuration inhibiting RNA unwinding compared with the unconstrained protein. Moreover, we found that differences in relative positioning of the cassettes strongly influence RNA-stimulated ATP hydrolysis by the N-terminal cassette. Our results indicate that the inactive C-terminal cassette of BRR2 can both positively and negatively affect the activity of the N-terminal helicase unit from a distance.
Keywords: ATPase; RNA helicase; RNA splicing; allosteric regulation; dual-cassette Ski2-like helicase; intramolecular regulation; nucleic acid-dependent nucleotide triphosphatase (NTPase); pre-mRNA splicing; protein conformation; small nuclear ribonucleoprotein U5 subunit 200 (SNRNP200); superfamily 2 helicase.
© 2020 Vester et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures



References
-
- Halls C., Mohr S., Del Campo M., Yang Q., Jankowsky E., and Lambowitz A. M. (2007) Involvement of DEAD-box proteins in group I and group II intron splicing: biochemical characterization of Mss116p, ATP hydrolysis-dependent and -independent mechanisms, and general RNA chaperone activity. J. Mol. Biol. 365, 835–855 10.1016/j.jmb.2006.09.083 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Research Materials

