Structure-guided development of selective M3 muscarinic acetylcholine receptor antagonists
- PMID: 30404914
- PMCID: PMC6255194
- DOI: 10.1073/pnas.1813988115
Structure-guided development of selective M3 muscarinic acetylcholine receptor antagonists
Abstract
Drugs that treat chronic obstructive pulmonary disease by antagonizing the M3 muscarinic acetylcholine receptor (M3R) have had a significant effect on health, but can suffer from their lack of selectivity against the M2R subtype, which modulates heart rate. Beginning with the crystal structures of M2R and M3R, we exploited a single amino acid difference in their orthosteric binding pockets using molecular docking and structure-based design. The resulting M3R antagonists had up to 100-fold selectivity over M2R in affinity and over 1,000-fold selectivity in vivo. The crystal structure of the M3R-selective antagonist in complex with M3R corresponded closely to the docking-predicted geometry, providing a template for further optimization.
Keywords: G protein-coupled receptor; crystal structure; drug design; muscarinic receptor; subtype selectivity.
Copyright © 2018 the Author(s). Published by PNAS.
Conflict of interest statement
Conflict of interest statement: B.K.K. is a cofounder of and consultant for ConfometRx, Inc.
Figures
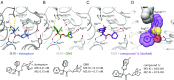
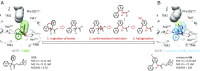

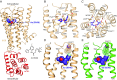
References
-
- Hulme EC, Birdsall NJM, Buckley NJ. Muscarinic receptor subtypes. Annu Rev Pharmacol Toxicol. 1990;30:633–673. - PubMed
-
- Wess J. Molecular biology of muscarinic acetylcholine receptors. Crit Rev Neurobiol. 1996;10:69–99. - PubMed
-
- Caulfield MP, Birdsall NJM. International Union of Pharmacology. XVII. Classification of muscarinic acetylcholine receptors. Pharmacol Rev. 1998;50:279–290. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

