Proteasome substrate capture and gate opening by the accessory factor PafE from Mycobacterium tuberculosis
- PMID: 29414791
- PMCID: PMC5880150
- DOI: 10.1074/jbc.RA117.001471
Proteasome substrate capture and gate opening by the accessory factor PafE from Mycobacterium tuberculosis
Abstract
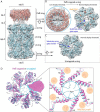
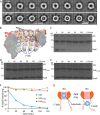
In all domains of life, proteasomes are gated, chambered proteases that require opening by activators to facilitate protein degradation. Twelve proteasome accessory factor E (PafE) monomers assemble into a single dodecameric ring that promotes proteolysis required for the full virulence of the human bacterial pathogen Mycobacterium tuberculosis Whereas the best characterized proteasome activators use ATP to deliver proteins into a proteasome, PafE does not require ATP. Here, to unravel the mechanism of PafE-mediated protein targeting and proteasome activation, we studied the interactions of PafE with native substrates, including a newly identified proteasome substrate, the ParA-like protein, Rv3213c, and with proteasome core particles. We characterized the function of a highly conserved feature in bacterial proteasome activator proteins: a glycine-glutamine-tyrosine-leucine (GQYL) motif at their C termini that is essential for stimulating proteolysis. Using cryo-electron microscopy (cryo-EM), we found that the GQYL motif of PafE interacts with specific residues in the α subunits of the proteasome core particle to trigger gate opening and degradation. Finally, we also found that PafE rings have 40-Å openings lined with hydrophobic residues that form a chamber for capturing substrates before they are degraded, suggesting PafE has a previously unrecognized chaperone activity. In summary, we have identified the interactions between PafE and the proteasome core particle that cause conformational changes leading to the opening of the proteasome gate and have uncovered a mechanism of PafE-mediated substrate degradation. Collectively, our results provide detailed insights into the mechanism of ATP-independent proteasome degradation in bacteria.
Keywords: Mycobacterium tuberculosis; Proteasome activator; cryo-electron microscopy; proteasome; protein degradation; protein targeting; structural biology.
© 2018 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures




References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

