Structures of human calpain-3 protease core with and without bound inhibitor reveal mechanisms of calpain activation
- PMID: 29382717
- PMCID: PMC5857979
- DOI: 10.1074/jbc.RA117.001097
Structures of human calpain-3 protease core with and without bound inhibitor reveal mechanisms of calpain activation
Abstract
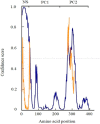
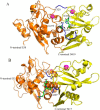
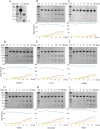
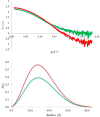
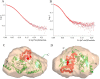
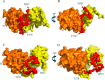
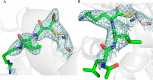
Limb-girdle muscular dystrophy type 2a arises from mutations in the Ca2+-activated intracellular cysteine protease calpain-3. This calpain isoform is abundant in skeletal muscle and differs from the main isoforms, calpain-1 and -2, in being a homodimer and having two short insertion sequences. The first of these, IS1, interrupts the protease core and must be cleaved for activation and substrate binding. Here, to learn how calpain-3 can be regulated and inhibited, we determined the structures of the calpain-3 protease core with IS1 present or proteolytically excised. To prevent intramolecular IS1 autoproteolysis, we converted the active-site Cys to Ala. Small-angle X-ray scattering (SAXS) analysis of the C129A mutant suggested that IS1 is disordered and mobile enough to occupy several locations. Surprisingly, this was also true for the apo version of this mutant. We therefore concluded that IS1 might have a binding partner in the sarcomere and is unstructured in its absence. After autoproteolytic IS1 removal from the active Cys129 calpain-3 protease core, we could solve its crystal structures with and without the cysteine protease inhibitors E-64 and leupeptin covalently bound to the active-site cysteine. In each structure, the active state of the protease core was assembled by the cooperative binding of two Ca2+ ions to the equivalent sites used in calpain-1 and -2. These structures of the calpain-3 active site with residual IS1 and with bound E-64 and leupeptin may help guide the design of calpain-3-specific inhibitors.
Keywords: autoproteolysis; calcium-binding protein; calpain-3; crystal structure; drug design; propeptide; protease inhibitor; protein complex; small-angle X-ray scattering (SAXS).
© 2018 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures



References
-
- Mellgren R. L., Miyake K., Kramerova I., Spencer M. J., Bourg N., Bartoli M., Richard I., Greer P. A., and McNeil P. L. (2009) Calcium-dependent plasma membrane repair requires m- or mu-calpain, but not calpain-3, the proteasome, or caspases. Biochim. Biophys. Acta. 1793, 1886–1893 10.1016/j.bbamcr.2009.09.013 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

