Chlamydial virulence factor TarP mimics talin to disrupt the talin-vinculin complex
- PMID: 29710402
- PMCID: PMC6001692
- DOI: 10.1002/1873-3468.13074
Chlamydial virulence factor TarP mimics talin to disrupt the talin-vinculin complex
Abstract
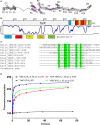
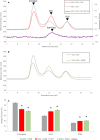
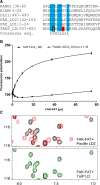
Vinculin is a central component of mechanosensitive adhesive complexes that form between cells and the extracellular matrix. A myriad of infectious agents mimic vinculin binding sites (VBS), enabling them to hijack the adhesion machinery and facilitate cellular entry. Here, we report the structural and biochemical characterisation of VBS from the chlamydial virulence factor TarP. Whilst the affinities of isolated VBS peptides from TarP and talin for vinculin are similar, their behaviour in larger fragments is markedly different. In talin, VBS are cryptic and require mechanical activation to bind vinculin, whereas the TarP VBS are located in disordered regions, and so are constitutively active. We demonstrate that the TarP VBS can uncouple talin:vinculin complexes, which may lead to adhesion destabilisation.
Keywords: adhesion; chlamydia; crystallography; molecular mimicry; talin; vinculin.
© 2018 The Authors. FEBS Letters published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Figures

References
-
- Izard T, Evans G, Borgon RA, Rush CL, Bricogne G and Bois PR (2004) Vinculin activation by talin through helical bundle conversion. Nature 427, 171–175. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

