Structure shows that the BIR2 domain of E3 ligase XIAP binds across the RIPK2 kinase dimer interface
- PMID: 37673444
- PMCID: PMC10485824
- DOI: 10.26508/lsa.202201784
Structure shows that the BIR2 domain of E3 ligase XIAP binds across the RIPK2 kinase dimer interface
Abstract
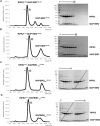
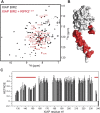
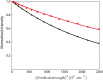
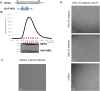
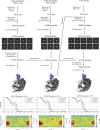
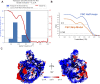
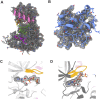
RIPK2 is an essential adaptor for NOD signalling and its kinase domain is a drug target for NOD-related diseases, such as inflammatory bowel disease. However, recent work indicates that the phosphorylation activity of RIPK2 is dispensable for signalling and that inhibitors of both RIPK2 activity and RIPK2 ubiquitination prevent the essential interaction between RIPK2 and the BIR2 domain of XIAP, the key RIPK2 ubiquitin E3 ligase. Moreover, XIAP BIR2 antagonists also block this interaction. To reveal the molecular mechanisms involved, we combined native mass spectrometry, NMR, and cryo-electron microscopy to determine the structure of the RIPK2 kinase BIR2 domain complex and validated the interface with in cellulo assays. The structure shows that BIR2 binds across the RIPK2 kinase antiparallel dimer and provides an explanation for both inhibitory mechanisms. It also highlights why phosphorylation of the kinase activation loop is dispensable for signalling while revealing the structural role of RIPK2-K209 residue in the RIPK2-XIAP BIR2 interaction. Our results clarify the features of the RIPK2 conformation essential for its role as a scaffold protein for ubiquitination.
© 2023 Lethier et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures









References
-
- Amar J, Chabo C, Waget A, Klopp P, Vachoux C, Bermúdez-Humarán LG, Smirnova N, Bergé M, Sulpice T, Lahtinen S, et al. (2011) Intestinal mucosal adherence and translocation of commensal bacteria at the early onset of type 2 diabetes: Molecular mechanisms and probiotic treatment. EMBO Mol Med 3: 559–572. 10.1002/emmm.201100159 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
