Drugging an undruggable pocket on KRAS
- PMID: 31332011
- PMCID: PMC6689897
- DOI: 10.1073/pnas.1904529116
Drugging an undruggable pocket on KRAS
Abstract
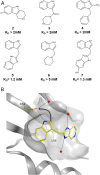
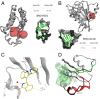
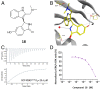
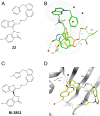
The 3 human RAS genes, KRAS, NRAS, and HRAS, encode 4 different RAS proteins which belong to the protein family of small GTPases that function as binary molecular switches involved in cell signaling. Activating mutations in RAS are among the most common oncogenic drivers in human cancers, with KRAS being the most frequently mutated oncogene. Although KRAS is an excellent drug discovery target for many cancers, and despite decades of research, no therapeutic agent directly targeting RAS has been clinically approved. Using structure-based drug design, we have discovered BI-2852 (1), a KRAS inhibitor that binds with nanomolar affinity to a pocket, thus far perceived to be "undruggable," between switch I and II on RAS; 1 is mechanistically distinct from covalent KRASG12C inhibitors because it binds to a different pocket present in both the active and inactive forms of KRAS. In doing so, it blocks all GEF, GAP, and effector interactions with KRAS, leading to inhibition of downstream signaling and an antiproliferative effect in the low micromolar range in KRAS mutant cells. These findings clearly demonstrate that this so-called switch I/II pocket is indeed druggable and provide the scientific community with a chemical probe that simultaneously targets the active and inactive forms of KRAS.
Keywords: KRAS; NMR; fragment-based drug design; oncology; structure-based drug design.
Copyright © 2019 the Author(s). Published by PNAS.
Conflict of interest statement
Conflict of interest statement: D.K., M.G., A.M., L.J.M., A.Z., M.M., A.G., D.C., S.F., T. Gerstberger, T. Gmashitz, P.G., D.H., W.H., J.H., J.K.-O., P.K., S.K., M.K., R.K., L.L., F.M., S.M.-M., C.P., J.R., C.S., Y.S., K.S., R.S., A.S., B.S., G.S., B.W., M.Z., M.P., and D.B.M. were employees of Boehringer Ingelheim at the time of this work.
Figures

Comment in
-
Researchers Reveal Another KRAS Inhibitor.Cancer Discov. 2019 Sep;9(9):1152. doi: 10.1158/2159-8290.CD-NB2019-092. Epub 2019 Aug 6. Cancer Discov. 2019. PMID: 31387866
-
Reply to Tran et al.: Dimeric KRAS protein-protein interaction stabilizers.Proc Natl Acad Sci U S A. 2020 Feb 18;117(7):3365-3367. doi: 10.1073/pnas.1921236117. Epub 2020 Feb 11. Proc Natl Acad Sci U S A. 2020. PMID: 32047042 Free PMC article. No abstract available.
-
The small molecule BI-2852 induces a nonfunctional dimer of KRAS.Proc Natl Acad Sci U S A. 2020 Feb 18;117(7):3363-3364. doi: 10.1073/pnas.1918164117. Epub 2020 Feb 11. Proc Natl Acad Sci U S A. 2020. PMID: 32047043 Free PMC article. No abstract available.
References
-
- Neumann J., Zeindl-Eberhart E., Kirchner T., Jung A., Frequency and type of KRAS mutations in routine diagnostic analysis of metastatic colorectal cancer. Pathol. Res. Pract. 205, 858–862 (2009). - PubMed
-
- Colombino M., et al. , BRAF/NRAS mutation frequencies among primary tumors and metastases in patients with melanoma. J. Clin. Oncol. 30, 2522–2529 (2012). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

