A SNARE geranylgeranyltransferase essential for the organization of the Golgi apparatus
- PMID: 32128853
- PMCID: PMC7156963
- DOI: 10.15252/embj.2019104120
A SNARE geranylgeranyltransferase essential for the organization of the Golgi apparatus
Abstract
Protein prenylation is essential for many cellular processes including signal transduction, cytoskeletal reorganization, and membrane trafficking. Here, we identify a novel type of protein prenyltransferase, which we named geranylgeranyltransferase type-III (GGTase-III). GGTase-III consists of prenyltransferase alpha subunit repeat containing 1 (PTAR1) and the β subunit of RabGGTase. Using a biotinylated geranylgeranyl analogue, we identified the Golgi SNARE protein Ykt6 as a substrate of GGTase-III. GGTase-III transfers a geranylgeranyl group to mono-farnesylated Ykt6, generating doubly prenylated Ykt6. The crystal structure of GGTase-III in complex with Ykt6 provides structural basis for Ykt6 double prenylation. In GGTase-III-deficient cells, Ykt6 remained in a singly prenylated form, and the Golgi SNARE complex assembly was severely impaired. Consequently, the Golgi apparatus was structurally disorganized, and intra-Golgi protein trafficking was delayed. Our findings reveal a fourth type of protein prenyltransferase that generates geranylgeranyl-farnesyl Ykt6. Double prenylation of Ykt6 is essential for the structural and functional organization of the Golgi apparatus.
Keywords: SNARE; Golgi; PTAR1; Ykt6; protein prenylation.
© 2020 The Authors.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

- A
PTAR1 protein expression in rat tissues. Proteins extracted from various rat tissues (15 μg each) were analyzed by immunoblotting with antibodies against PTAR1.
- B
Identification of RabGGTβ as a binding protein of PTAR1. Anti‐FLAG immunoprecipitates from control HeLa S3 cells or FLAG‐PTAR1‐expressing HeLa S3 cells were analyzed by SDS–PAGE and silver staining. The 35 kDa protein was identified as RabGGTβ by mass spectrometry. The asterisk denotes a degradation product of FLAG‐PTAR1.
- C
Co‐immunoprecipitation of endogenous PTAR1 and RabGGTβ from rat brain and thymus cytosol.
- D
Fractionation of rat brain cytosol by Mono Q anion exchange column chromatography. Column fractions were analyzed by immunoblotting with antibodies against RabGGTα, PTAR1, and RabGGTβ. RabGGTβ eluted in two peaks (peak A and peak B).
- E
Co‐immunodepletion of PTAR1 and RabGGTβ by anti‐PTAR1 IgG. PTAR1 was immunodepleted from the Mono Q peak A fraction using anti‐PTAR1 IgG‐bound protein A beads, and the supernatant was analyzed by immunoblotting with antibodies against PTAR1 and RabGGTβ. Normal rabbit IgG and anti‐PTAR1 IgG preabsorbed with the antigen peptide were used as controls.
- F
Fractionation of the Mono Q peak A fraction by Superdex 200 gel filtration chromatography. Column fractions were analyzed by immunoblotting with antibodies against PTAR1 and RabGGTβ.
- G
Gel filtration chromatography of the recombinant PTAR1–RabGGTβ complex purified from Sf9 insect cells on a Superdex 200 column. Inset shows SDS–PAGE and Coomassie staining analysis of the purified complex.
- H
Schematic diagram of the protein prenyltransferase family. FTase and GGTase‐I share the same α subunit. RabGGTase and the PTAR1–RabGGTβ complex (GGTase‐III) share the same β subunit.

- A–D
Prenylation activity of GGTase‐III on Ras, Rho, and Rab proteins. (A) His6‐H‐Ras (5 μM) was incubated with FTase (50 nM) or GGTase‐III (50 nM) and 3H‐FPP (1 μM) at 37°C. Reactions were stopped at the indicated time points, and the amount of 3H‐farnesyl transferred to H‐Ras was quantified by scintillation counting (mean ± SEM, n = 3). (B) His6‐RhoA (5 μM) was incubated with GGTase‐I (50 nM) or GGTase‐III (50 nM) and 3H‐GGPP (1 μM), and the amount of 3H‐geranylgeranyl transferred to RhoA was quantified (mean ± SEM, n = 3). (C) His6‐Rab1A (5 μM) was incubated with RabGGTase (50 nM) or GGTase‐III (50 nM) and 3H‐GGPP (1 μM) in the absence or presence of either His6‐REP1 (100 nM) or His6‐REP2 (100 nM), and the amount of 3H‐geranylgeranyl transferred to Rab1A was quantified (mean ± SEM, n = 3). (D) His6‐Rab5A and His6‐Rab11A were incubated with RabGGTase (50 nM) or GGTase‐III (50 nM) and 3H‐GGPP (1 μM) in the presence of His6‐REP1 (100 nM), and the amount of 3H‐geranylgeranyl transferred to Rab5A and Rab11A was quantified (mean ± SEM, n = 3).
- E
REP pull‐down assay. Recombinant His6‐REP1 or His6‐REP2 was incubated with glutathione Sepharose beads coated with GST, GST‐RabGGTase, or GST‐GGTase‐III. Bound His6‐REP proteins were analyzed by immunoblotting with anti‐His6 antibody.
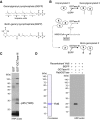
- A
Molecular structures of geranylgeranyl pyrophosphate (GGPP) and its biotinylated analogue biotin‐geranyl pyrophosphate (BGPP). Geranylgeranyl moiety consists of four repeats of 5‐carbon isoprene unit.
- B
Purification strategy for GGTase‐III substrate proteins using statin and BGPP. “X” represents a putative substrate protein of GGTase‐III. HMG‐CoA, hydroxymethylglutaryl‐CoA.
- C
Identification of Ykt6 as a protein biotin‐geranylated by GGTase‐III. Cytosolic proteins extracted from statin‐treated HeLa S3 cells were applied to GST or GST‐GGTase‐III affinity columns. Bound proteins were eluted and incubated with recombinant GGTase‐III and BGPP. Reaction products were separated by SDS–PAGE, transferred to a nitrocellulose membrane, and probed with horseradish peroxidase (HRP)‐labeled avidin to detect biotinylated proteins. Mass spectrometry identified the 25 kDa protein as Ykt6.
- D
Biotin‐geranylation of recombinant Ykt6 by GGTase‐III. Bacterially produced recombinant untagged Ykt6 (left) was incubated with buffer, GGTase‐III, or RabGGTase in the absence or presence of BGPP for 30 min at 37°C. Biotin‐geranylated Ykt6 was detected as in (C).

- A
Schematic diagram showing the domain structure of human Ykt6 protein. Ykt6 has two cysteine residues, Cys194 and Cys195, at the fifth and fourth positions from the C‐terminus.
- B
Alignment of the C‐terminal amino acid sequences of Ykt6 from various species. The conserved tandem cysteines are highlighted in gray.
- C
Farnesylation of recombinant Ykt6 by FTase. Recombinant Ykt6 (5 μM) was incubated with FTase (0 or 50 nM) and 3H‐FPP (1 μM; ˜ 3,500 dpm/pmol) at 37°C. Reactions were stopped at the indicated time points, and the amount of 3H‐farnesyl transferred to Ykt6 was quantified by scintillation counting (mean ± SEM, n = 3).
- D
Geranylgeranylation of recombinant Ykt6 by GGTase‐III. Recombinant Ykt6 (5 μM) was incubated with increasing concentrations of GGTase‐III (0–100 nM) and 3H‐GGPP (1 μM; ˜ 3,000 dpm/pmol) for the indicated times, and the amount of 3H‐geranylgeranyl transferred to Ykt6 was quantified (mean ± SEM, n = 3).
- E
Geranylgeranylation of Cys to Ser mutants of Ykt6 by GGTase‐III. WT and mutant Ykt6 proteins (5 μM each) were incubated with GGTase‐III (100 nM) and 3H‐GGPP (1 μM) for the indicated times, and the amount of 3H‐geranylgeranyl transferred to Ykt6 was quantified (mean ± SEM, n = 3).
- F
Sequential prenylation process of Ykt6. SAM, S‐adenosylmethionine; SAH, S‐adenosylhomocysteine; PPi, inorganic pyrophosphate.
- G
Mass spectra of prenylated Ykt6 peptides. Recombinant Ykt6 was farnesylated by FTase (FTase), further processed by RCE1 and ICMT (RCE1/ICMT), and finally geranylgeranylated by GGTase‐III (GGTase‐III). After purification to homogeneity, the prenylated Ykt6 proteins were digested with chymotrypsin (left) or V8 protease (right) and analyzed by MALDI‐TOF mass spectrometry. The calculated m/z values for the C‐terminal peptide ions are shown in parentheses. The peaks labeled with an asterisk correspond to amino acids 14–38 of Ykt6 (AKVVLLKAAYDVSSFSFFQRSSVQE; m/z 2,806.48).
- H
Effects of C‐terminal processing and Cys195 farnesylation on Cys194 geranylgeranylation by GGTase‐III. Purified recombinant unprenyl Ykt6, Cys195‐farnesyl Ykt6, and Cys195‐farnesyl Ykt6ΔAIM (5 μM each) were incubated with GGTase‐III (100 nM) and 3H‐GGPP (1 μM) for the indicated times, and the amount of 3H‐geranylgeranyl transferred to Ykt6 was quantified (mean ± SEM, n = 3).
- I
Gel filtration chromatography of purified unprenyl Ykt6, Cys195‐farnesyl Ykt6ΔAIM, and Cys194/195‐diprenyl Ykt6ΔAIM (135 μg each) on a Superdex 200 column. Dotted lines indicate the elution profile of unprenyl Ykt6 for comparison. The prenylated Ykt6 proteins were eluted slightly later than the unprenylated Ykt6, suggesting more compact and stable folds.

- A
Structural comparison of GGTase‐III and other prenyltransferases. The α and β subunits of the prenyltransferases are shown in green and cyan, respectively. The unique N‐terminal domain of PTAR1 is shown in yellow.
- B
Overall structure of GGTase‐III complexed with Cys195‐farnesyl Ykt6ΔAIM and GGPP in two orientations. GGTase‐III is colored as in (A). The longin domain of Ykt6 is in pink, and SNARE domain is in magenta.
- C, D
Magnified views of site 1 (C) and site 2 (D) depicting the interaction between PTAR1 and Ykt6. Dashed lines indicate hydrogen bonds.
- E
Geranylgeranylation activity of WT GGTase‐III and the indicated mutants (100 nM each). Cys195‐farnesyl Ykt6ΔAIM (1 μM) and 3H‐GGPP (1 μM) were used as substrates (mean ± SEM, n = 3).
- F
Kinetic analysis of WT GGTase‐III and the site 1 or site 2 mutant (100 nM each) using increasing concentrations of Cys195‐farnesyl Ykt6ΔAIM (mean ± SEM, n = 3 for WT and Y306A, n = 1 for V48A/V50A).
- G
Geranylgeranylation of unprenyl WT Ykt6 and the site 1 or site 2 mutants by GGTase‐III. WT and mutant Ykt6 proteins (5 μM each) were incubated with GGTase‐III (100 nM) and 3H‐GGPP (1 μM), and the amount of 3H‐geranylgeranyl transferred to Ykt6 was quantified (mean ± SEM, n = 3).

- A
Overall view of the central cavity of the β subunit showing insertion of the C‐terminal tail of Ykt6. PTAR1 is omitted for clarity. The Cys194‐linked geranylgeranyl and Cys195‐linked farnesyl moieties are shown in orange and purple, respectively. PPi, inorganic pyrophosphate.
- B
Electron density of the Cys194‐linked geranylgeranyl and Cys195‐linked farnesyl moieties (blue mesh; simulated‐annealing F o–F c omit map contoured at 3σ level).
- C, D
Detailed views of the geranylgeranyl moiety in the GGTase‐III–product complex (C) and GGPP in the GGTase‐III–GGPP complex (D). Residues of the β subunit that form the lipid substrate binding pocket are shown.
- E
Surface representation of the central cavity of the β subunit showing a tunnel formed near the active site. The Cys195‐linked farnesyl moiety is anchored into the hydrophobic tunnel.
- F
Detailed view of the Cys195‐linked farnesyl group bound in the hydrophobic tunnel. Residues of the β subunit that line the hydrophobic tunnel are shown.
- G
Superposition of the β subunits of FTase (blue), GGTase‐I (gray), and GGTase‐III (cyan). In FTase and GGTase‐I, the hydrophobic tunnel is blocked by the side chain of Leu103 and Ile50, respectively. The corresponding position is replaced by Gly49 in GGTase‐III.
- H
Geranylgeranylation activity of WT GGTase‐III and tunnel mutants (100 nM each). Cys195‐farnesyl Ykt6ΔAIM (1 μM) and 3H‐GGPP (1 μM) were used as substrates (mean ± SEM, n = 3).
- I
GGPP binding to GGTase‐III. WT GGTase‐III and tunnel mutants (1 μM each) were incubated with 3H‐GGPP (2 μM) at 4°C for 10 min. After desalting, enzyme‐bound 3H‐GGPP was quantified by scintillation counting (mean ± SEM, n = 3).
- J
Alignment of the helix 3 sequences of RabGGTβ orthologues and human FTβ and GGT‐Iβ. The position of the conserved glycine residue (Gly49 in human) is highlighted in gray.
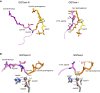
- A
Left, comparison of the Cys194‐linked geranylgeranyl moiety (orange) in the GGTase‐III–product complex and GGPP (yellow) in the GGTase‐III–GGPP complex. In the product complex, the first and second isoprene units of GGPP are rotated to form a covalent bond with Cys194. The Cys195‐linked farnesyl moiety (purple) is anchored into the hydrophobic tunnel. Right, comparison of the geranylgeranyl moiety (orange) linked to Cys4 of C4VIL peptide in the GGTase‐I–product complex (PDB 1N4R) and GGPP (yellow) in the GGTase‐I–GGPP complex (PDB 1N4P). GGTase‐III and GGTase‐I show similar isoprene movements during catalysis.
- B
Left, the catalytic zinc ion and zinc‐coordinating residues of RabGGTase (gray; PDB 3DST) superposed on the GGTase‐III–product complex (cyan). The sulfur atom of geranylgeranylated Cys194 is located in the vicinity of the superposed catalytic zinc ion. Right, the catalytic site of the GGTase‐I–product complex (PDB 1N4R). GGTase‐III has a similar active site arrangement as GGTase‐I.

- A
Generation of PTAR1 KO cell lines by CRISPR‐Cas9 using single‐guide RNA (sgRNA) targeting exon 2 of the human PTAR1 gene. The target sequence and PAM sequence are indicated.
- B
Sequence data of the genomic region containing the exon 2 of PTAR1. Insertion and deletion mutations are shown in red, and the resulting premature stop codons are underlined. These mutations introduce a stop codon at the amino acid position 42 or 43. HAP1 is a haploid cell line and contains a single allele for PTAR1. nt, nucleotide.
- C
Immunoblot analysis of WT cells, PTAR1 KO cells, and PTAR1 KO cells stably expressing PTAR1 (KO + PTAR1) using anti‐PTAR1 antibody. Asterisks denote proteins that cross‐react with the antibody.
- D
Cytosolic localization of Ykt6 in HeLa cells. WT and PTAR1 KO HeLa cells were fractionated into cytosol and membrane fractions, and analyzed by immunoblotting with anti‐syntaxin 5 and anti‐Ykt6 antibody. Syntaxin 5 has two isoforms with different translation initiation sites.
- E
Prenylation status of Ykt6 in PTAR1 KO HAP1 cells and PTAR1 KO HAP1 cells stably expressing WT PTAR1 or Yk6‐binding defective mutants of PTAR1. The prenylation status of Ykt6 was analyzed by DOC‐PAGE followed by immunoblotting with anti‐Ykt6 antibody. Lower panels show conventional immunoblot analysis of the same samples using the indicated antibodies.

- A
Immunoblot of cell lysates from WT HAP1 cells and PTAR1 KO HAP1 cells using anti‐Ykt6 antibody. Note the slight difference in the gel mobility of Ykt6 (arrows).
- B
Prenylation status of rat brain Ykt6. Recombinant unprenyl Ykt6, Cys195‐farnesyl Ykt6, Cys194/195‐diprenyl Ykt6 samples (1 ng each), and rat brain cytosol were electrophoresed on a polyacrylamide gel using deoxycholate (DOC)‐containing buffer and analyzed by immunoblotting with anti‐Ykt6 antibody (DOC‐PAGE; upper panel). The same samples were analyzed by conventional SDS–PAGE and immunoblotting with anti‐Ykt6 antibody (SDS–PAGE; lower panel).
- C
Prenylation status of Ykt6 in WT and PTAR1 KO cells. Recombinant Ykt6 samples and cell lysates of WT cells, PTAR1 KO cells, and PTAR1 KO cells stably expressing PTAR1 (KO + PTAR1) were separated by DOC‐PAGE (upper) or SDS–PAGE (lower), and analyzed by immunoblotting with anti‐Ykt6 antibody.
- D
In vitro reconstitution of Ykt6 double prenylation. Dialyzed cytosol prepared from PTAR1 KO HAP1 cells was incubated at 37°C for the indicated times with recombinant GGTase‐III (100 nM) or RabGGTase (100 nM) in the absence or presence of GGPP (10 μM). After incubation, reaction products were separated by DOC‐PAGE (upper) or SDS–PAGE (lower), and analyzed by immunoblotting with anti‐Ykt6 antibody.
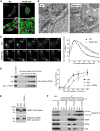
- A
Representative confocal images of the Golgi apparatus in WT or PTAR1 KO HeLa cells. Cells were co‐immunostained for GM130 (a cis‐Golgi marker) and golgin‐97 (a trans‐Golgi marker). Images were deconvoluted using Huygens software. The lower panels show magnified images of the boxed region of the upper panels. Scale bars, 5 μm.
- B
Electron micrographs of the Golgi apparatus in WT or PTAR1 KO HAP1 cells. Arrows indicate Golgi stacks. Arrowheads indicate examples of unfused vesicles accumulated around the swollen Golgi cisternae in PTAR1 KO cells. Scale bars, 500 nm.
- C, D
Defect in intra‐Golgi trafficking in PTAR1 KO cells. (C) VSVG‐GFP expressing WT and PTAR1 KO HeLa cells were cultured at 40°C and then shifted to 32°C. VSVG‐GFP fluorescence images taken at the indicated times after the temperature shift are shown. The right panel shows quantification of VSVG‐GFP fluorescence in the Golgi region after the temperature shift (mean ± SEM, n = 6). (D) VSVG‐GFP expressing WT HeLa cells, PTAR1 KO HeLa cells, and PTAR1 KO HeLa cells stably expressing PTAR1 (KO + PTAR1) were cultured at 40°C and then shifted to 32°C. At the indicated time points, cell surface proteins were biotinylated using sulfo‐NHS‐LC‐biotin. Biotinylated VSVG‐GFP was purified from cell lysates using avidin agarose and analyzed by immunoblotting with anti‐GFP antibody. The right panel shows quantification of the cell surface biotinylated VSVG‐GFP (means ± SEM, n = 3). Data were analyzed by one‐way ANOVA with Dunnett's post‐hoc test. *P < 0.05, ***P < 0.001.
- E
Analysis of LAMP1 glycosylation. Cell lysates of WT HeLa cells, PTAR1 KO HeLa cells, and PTAR1 KO HeLa cells stably expressing PTAR1 (KO + PTAR1) were analyzed by immunoblotting with anti‐LAMP1 antibody (upper). Sialylated LAMP1 was precipitated from the cell lysates using Maackia amurensis leucoagglutinin (MAL) agarose and analyzed by immunoblotting (lower).
- F
Defect in the Golgi SNARE assembly in PTAR1 KO cells. The Golgi SNARE complex was immunoprecipitated from NEM‐treated WT HAP1 cells, PTAR1 KO HAP1 cells, and PTAR1 KO HAP1 cells stably expressing PTAR1 (KO + PTAR1) using control mouse IgG, anti‐GS28 IgG, or anti‐syntaxin 5 IgG. Immunoprecipitates were analyzed by immunoblotting with antibodies against syntaxin 5, GS28, GS15, and Ykt6. Syntaxin 5 has two isoforms with different translation initiation sites. Inputs were 20% (syntaxin 5, GS28, and GS15) and 2% (Ykt6). The data shown are representative of three independent experiments with similar results.

- A
Model for the sequential double prenylation of Ykt6. (1) Nascent unprenylated Ykt6 exists in both closed and open conformations. (2) Farnesylation of Cys195 stabilizes the closed conformation of Ykt6. (3) Doubly prenylated Ykt6 keeps the closed conformation by sequestering both farnesyl and geranylgeranyl groups into the putative prenyl binding groove. (4) Upon activation, the SNARE domain is unfolded and the C‐terminal two prenyl groups are inserted into the membrane.
- B
Superposition of the dodecylphosphocholine (DPC)‐bound, closed form of Ykt6 (gray; PDB 3KYQ) and Cys194/195‐diprenyl Ykt6ΔAIM complexed to GGTase‐III (longin, pink; SNARE, magenta). GGTase‐III is omitted for clarity. The putative prenyl binding groove of Ykt6, occupied by DPC (yellow), is located closed to the active site of the enzyme. Upon binding to GGTase‐III, the Cys195‐linked farnesyl moiety accommodated in the putative prenyl binding groove of Ykt6 is translocated into the hydrophobic tunnel of the enzyme, allowing the transfer of geranylgeranyl moiety to Cys194. The attached two prenyl groups may easily translocate back to the prenyl binding groove of Ykt6.
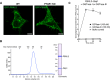
- A
Localization of FBXL2 in HeLa cells. WT HeLa cells and PTAR1 KO HeLa cells transiently expressing GFP‐FBXL2 and myc‐Skp1 were observed for GFP fluorescence. In PTAR1 KO cells, GFP‐FBXL2 was still able to localize to the plasma membrane as observed in WT cells. Scale bars, 10 μm.
- B
Purification of the recombinant FBXL2–Skp1 complex by Superdex 200 gel filtration chromatography. The peak fraction was analyzed by SDS–PAGE and Coomassie staining.
- C
Geranylgeranylation assay showing that GGTase‐III cannot geranylgeranylate FBXL2. The purified recombinant FBXL2–Skp1 complex (5 μM) was incubated with GGTase‐I (100 nM) or GGTase‐III (100 nM) and 3H‐GGPP (1 μM) at 37°C. Reactions were stopped at the indicated time points, and the amount of 3H‐geranylgeranyl transferred to FBXL2 was quantified by scintillation counting (mean ± SEM, n = 3). GGTase‐III failed to geranylgeranylate FBXL2, whereas GGTase‐I efficiently geranylgeranylated FBXL2 under the same conditions.
Comment in
-
Geranylgeranyl generosity: a new prenyl-transferase gives a fat to a SNARE protein.EMBO J. 2020 Apr 15;39(8):e104744. doi: 10.15252/embj.2020104744. Epub 2020 Mar 23. EMBO J. 2020. PMID: 32202660 Free PMC article.
References
-
- Andres DA, Seabra MC, Brown MS, Armstrong SA, Smeland TE, Cremers FP, Goldstein JL (1993) cDNA cloning of component A of Rab geranylgeranyl transferase and demonstration of its role as a Rab escort protein. Cell 73: 1091–1099 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
- 16K08574/MEXT | Japan Society for the Promotion of Science (JSPS)/International
- 17K15072/MEXT | Japan Society for the Promotion of Science (JSPS)/International
- 16H05148/MEXT | Japan Society for the Promotion of Science (JSPS)/International
- Takeda Science Foundation/International
- JPMJCR12M5/MEXT | JST | Core Research for Evolutional Science and Technology (CREST)/International
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

