Structural basis for the specific cleavage of core-fucosylated N-glycans by endo-β- N-acetylglucosaminidase from the fungus Cordyceps militaris
- PMID: 31548313
- PMCID: PMC6851319
- DOI: 10.1074/jbc.RA119.010842
Structural basis for the specific cleavage of core-fucosylated N-glycans by endo-β- N-acetylglucosaminidase from the fungus Cordyceps militaris
Abstract
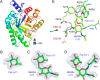
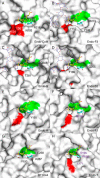
N-Linked glycans play important roles in various cellular and immunological events. Endo-β-N-acetylglucosaminidase (ENGase) can release or transglycosylate N-glycans and is a promising tool for the chemoenzymatic synthesis of glycoproteins with homogeneously modified glycans. The ability of ENGases to act on core-fucosylated glycans is a key factor determining their therapeutic utility because mammalian N-glycans are frequently α-1,6-fucosylated. Although the biochemistries and structures of various ENGases have been studied extensively, the structural basis for the recognition of the core fucose and the asparagine-linked GlcNAc is unclear. Herein, we determined the crystal structures of a core fucose-specific ENGase from the caterpillar fungus Cordyceps militaris (Endo-CoM), which belongs to glycoside hydrolase family 18. Structures complexed with fucose-containing ligands were determined at 1.75-2.35 Å resolutions. The fucose moiety linked to GlcNAc is extensively recognized by protein residues in a round-shaped pocket, whereas the asparagine moiety linked to the GlcNAc is exposed to the solvent. The N-glycan-binding cleft of Endo-CoM is Y-shaped, and several lysine and arginine residues are present at its terminal regions. These structural features were consistent with the activity of Endo-CoM on fucose-containing glycans on rituximab (IgG) and its preference for a sialobiantennary substrate. Comparisons with other ENGases provided structural insights into their core fucose tolerance and specificity. In particular, Endo-F3, a known core fucose-specific ENGase, has a similar fucose-binding pocket, but the surrounding residues are not shared with Endo-CoM. Our study provides a foothold for protein engineering to develop enzymatic tools for the preparation of more effective therapeutic antibodies.
Keywords: Cordyceps militaris; GH18; N-linked glycosylation; X-ray crystallography; core-fucosylated N-glycan; endo-β-N-acetylglucosaminidase (ENGase); glycoprotein; glycoside hydrolase; monoclonal antibody; substrate-assisted mechanism.
© 2019 Seki et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures


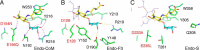

References
-
- Bieberich E. (2014) in Glycobiology of the Nervous System (Yu R. K., and Schengrund C.-L., eds) pp. 47–70, Springer, New York: 10.1007/978-1-4939-1154-7_3 - DOI
-
- Stanley P., Taniguchi N., and Aebi M. (2017) Essentials of Glycobiology, 3rd Ed, Chapter 9, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: 10.1101/glycobiology.3e.009 - DOI
-
- Pučić M., Knežević A., Vidič J., Adamczyk B., Novokmet M., Polašek O., Gornik O., Šupraha Goreta S., Wormald M. R., Redžic I., Campbell H., Wright A., Hastie N. D., Wilson J. F., Rudan I., et al. (2011) High throughput isolation and glycosylation analysis of IgG-variability and heritability of the IgG glycome in three isolated human populations. Mol. Cell. Proteomics 10, M111.010090 10.1074/mcp.M111.010090 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

