High affinity binding of SARS-CoV-2 spike protein enhances ACE2 carboxypeptidase activity
- PMID: 33122196
- PMCID: PMC7833600
- DOI: 10.1074/jbc.RA120.015303
High affinity binding of SARS-CoV-2 spike protein enhances ACE2 carboxypeptidase activity
Abstract
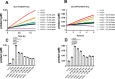
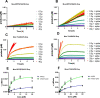
The novel severe acute respiratory syndrome coronavirus (SARS-CoV-2) has emerged to a pandemic and caused global public health crisis. Human angiotensin-converting enzyme 2(ACE2) was identified as the entry receptor for SARS-CoV-2. As a carboxypeptidase, ACE2 cleaves many biological substrates besides angiotensin II to control vasodilatation and vascular permeability. Given the nanomolar high affinity between ACE2 and SARS-CoV-2 spike protein, we investigated how this interaction would affect the enzymatic activity of ACE2. Surprisingly, SARS-CoV-2 trimeric spike protein increased ACE2 proteolytic activity ∼3-10 fold against model peptide substrates, such as caspase-1 substrate and Bradykinin-analog. The enhancement in ACE2 enzymatic function was mediated by the binding of SARS-CoV-2 spike RBD domain. These results highlighted the potential for SARS-CoV-2 infection to enhance ACE2 activity, which may be relevant to the cardiovascular symptoms associated with COVID-19.
Keywords: SARS-CoV-2 spike protein; angiotensin converting enzyme 2; carboxypeptidase; enzymatic activity; fluorescence resonance energy transfer (FRET); pathogenesis; renin angiotensin system; viral protein.
Conflict of interest statement
Conflict of interest—The authors declare that they have no conflicts of interest with the contents f this article.
Figures




Update of
-
High affinity binding of SARS-CoV-2 spike protein enhances ACE2 carboxypeptidase activity.bioRxiv [Preprint]. 2020 Jul 1:2020.07.01.182659. doi: 10.1101/2020.07.01.182659. bioRxiv. 2020. Update in: J Biol Chem. 2020 Dec 25;295(52):18579-18588. doi: 10.1074/jbc.RA120.015303. PMID: 32637947 Free PMC article. Updated. Preprint.
References
-
- Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., Yuan M.-L., Zhang Y.-L., Dai F.-H., Liu Y., Wang Q.-M. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. 32015508. - DOI - PMC - PubMed
-
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. 32015507. - DOI - PMC - PubMed
-
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., China Novel Coronavirus Investigating and Research Team A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. 31978945. - DOI - PMC - PubMed
-
- Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., Liu S., Zhao P., Liu H., Zhu L., Tai Y., Bai C., Gao T., Song J., Xia P. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

