Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency
- PMID: 32284326
- PMCID: PMC7242698
- DOI: 10.1074/jbc.RA120.013679
Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency
Abstract
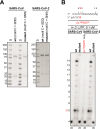
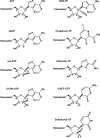
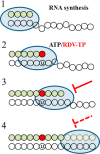
Effective treatments for coronavirus disease 2019 (COVID-19) are urgently needed to control this current pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Replication of SARS-CoV-2 depends on the viral RNA-dependent RNA polymerase (RdRp), which is the likely target of the investigational nucleotide analogue remdesivir (RDV). RDV shows broad-spectrum antiviral activity against RNA viruses, and previous studies with RdRps from Ebola virus and Middle East respiratory syndrome coronavirus (MERS-CoV) have revealed that delayed chain termination is RDV's plausible mechanism of action. Here, we expressed and purified active SARS-CoV-2 RdRp composed of the nonstructural proteins nsp8 and nsp12. Enzyme kinetics indicated that this RdRp efficiently incorporates the active triphosphate form of RDV (RDV-TP) into RNA. Incorporation of RDV-TP at position i caused termination of RNA synthesis at position i+3. We obtained almost identical results with SARS-CoV, MERS-CoV, and SARS-CoV-2 RdRps. A unique property of RDV-TP is its high selectivity over incorporation of its natural nucleotide counterpart ATP. In this regard, the triphosphate forms of 2'-C-methylated compounds, including sofosbuvir, approved for the management of hepatitis C virus infection, and the broad-acting antivirals favipiravir and ribavirin, exhibited significant deficits. Furthermore, we provide evidence for the target specificity of RDV, as RDV-TP was less efficiently incorporated by the distantly related Lassa virus RdRp, and termination of RNA synthesis was not observed. These results collectively provide a unifying, refined mechanism of RDV-mediated RNA synthesis inhibition in coronaviruses and define this nucleotide analogue as a direct-acting antiviral.
Keywords: COVID-19; Ebola virus; Lassa virus; MERS; RNA polymerase; RNA-dependent RNA polymerase (RdRp); SARS; SARS-CoV-2; coronavirus (CoV); drug action; drug development; drug discovery; favipiravir; plus-stranded RNA virus; remdesivir; replication; ribavirin; sofosbuvir.
© 2020 Gordon et al.
Conflict of interest statement
M. G. has previously received funding from Gilead Sciences in support of the study of EBOV RdRp inhibition by RDV. This study is also sponsored in part by a grant from Gilead Sciences to M. G., J. K. P., J. Y. F., and D. P. P. are Gilead employees
Figures



References
-
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., et al. (2020) Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395, 565–574 10.1016/S0140-6736(20)30251-8 - DOI - PMC - PubMed
-
- World Health Organization (2020) Coronavirus Disease 2019 (COVID-19) Situation Report-50, March 10, World Health Organization, Geneva
-
- Siegel D., Hui H. C., Doerffler E., Clarke M. O., Chun K., Zhang L., Neville S., Carra E., Lew W., Ross B., Wang Q., Wolfe L., Jordan R., Soloveva V., Knox J., et al. (2017) Discovery and synthesis of a phosphoramidate prodrug of a pyrrolo[2,1-f][triazin-4-amino] adenine C-nucleoside (GS-5734) for the treatment of Ebola and emerging viruses. J. Med. Chem. 60, 1648–1661 10.1021/acs.jmedchem.6b01594 - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

