Structural and functional characterization of the mitochondrial complex IV assembly factor Coa6
- PMID: 31515291
- PMCID: PMC6743065
- DOI: 10.26508/lsa.201900458
Structural and functional characterization of the mitochondrial complex IV assembly factor Coa6
Abstract
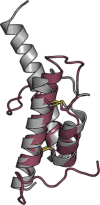
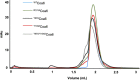
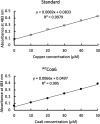
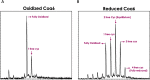
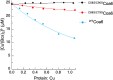
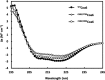
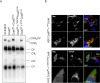
Assembly factors play key roles in the biogenesis of many multi-subunit protein complexes regulating their stability, activity, and the incorporation of essential cofactors. The human assembly factor Coa6 participates in the biogenesis of the CuA site in complex IV (cytochrome c oxidase, COX). Patients with mutations in Coa6 suffer from mitochondrial disease due to complex IV deficiency. Here, we present the crystal structures of human Coa6 and the pathogenic W59CCoa6-mutant protein. These structures show that Coa6 has a 3-helical bundle structure, with the first 2 helices tethered by disulfide bonds, one of which likely provides the copper-binding site. Disulfide-mediated oligomerization of the W59CCoa6 protein provides a structural explanation for the loss-of-function mutation.
© 2019 Maghool et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
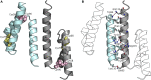




References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
