Burkholderia cenocepacia epigenetic regulator M.BceJIV simultaneously engages two DNA recognition sequences for methylation
- PMID: 39244607
- PMCID: PMC11380664
- DOI: 10.1038/s41467-024-52130-x
Burkholderia cenocepacia epigenetic regulator M.BceJIV simultaneously engages two DNA recognition sequences for methylation
Abstract
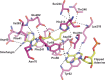
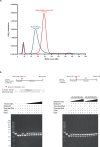
Burkholderia cenocepacia is an opportunistic and infective bacterium containing an orphan DNA methyltransferase called M.BceJIV with roles in regulating gene expression and motility of the bacterium. M.BceJIV recognizes a GTWWAC motif (where W can be an adenine or a thymine) and methylates N6 of the adenine at the fifth base position. Here, we present crystal structures of M.BceJIV/DNA/sinefungin ternary complex and allied biochemical, computational, and thermodynamic analyses. Remarkably, the structures show not one, but two DNA substrates bound to the M.BceJIV dimer, with each monomer contributing to the recognition of two recognition sequences. We also show that methylation at the two recognition sequences occurs independently, and that the GTWWAC motifs are enriched in intergenic regions in the genomes of B. cenocepacia strains. We further computationally assess the interactions underlying the affinities of different ligands (SAM, SAH, and sinefungin) for M.BceJIV, as a step towards developing selective inhibitors for limiting B. cenocepacia infection.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





Update of
-
Burkholderia cenocepacia epigenetic regulator M.BceJIV simultaneously engages two DNA recognition sequences for methylation.bioRxiv [Preprint]. 2024 Jan 23:2024.01.20.576384. doi: 10.1101/2024.01.20.576384. bioRxiv. 2024. Update in: Nat Commun. 2024 Sep 7;15(1):7839. doi: 10.1038/s41467-024-52130-x. PMID: 38328099 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

