Structure of the Complex of F-Actin and DNGR-1, a C-Type Lectin Receptor Involved in Dendritic Cell Cross-Presentation of Dead Cell-Associated Antigens
- PMID: 25979418
- PMCID: PMC5066845
- DOI: 10.1016/j.immuni.2015.04.009
Structure of the Complex of F-Actin and DNGR-1, a C-Type Lectin Receptor Involved in Dendritic Cell Cross-Presentation of Dead Cell-Associated Antigens
Abstract
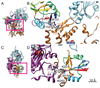
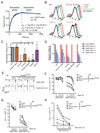
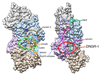
DNGR-1 is a C-type lectin receptor that binds F-actin exposed by dying cells and facilitates cross-presentation of dead cell-associated antigens by dendritic cells. Here we present the structure of DNGR-1 bound to F-actin at 7.7 Å resolution. Unusually for F-actin binding proteins, the DNGR-1 ligand binding domain contacts three actin subunits helically arranged in the actin filament, bridging over two protofilaments, as well as two neighboring actin subunits along one protofilament. Mutation of residues predicted to mediate ligand binding led to loss of DNGR-1-dependent cross-presentation of dead cell-associated antigens, formally demonstrating that the latter depends on F-actin recognition. Notably, DNGR-1 has relatively modest affinity for F-actin but multivalent interactions allow a marked increase in binding strength. Our findings shed light on modes of actin binding by cellular proteins and reveal how extracellular detection of cytoskeletal components by dedicated receptors allows immune monitoring of loss of cellular integrity.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures




References
-
- Abdiche Y, Malashock D, Pinkerton A, Pons J. Determining kinetics and affinities of protein interactions using a parallel real-time label-free biosensor, the Octet. Analytical biochemistry. 2008;377:209–217. - PubMed
-
- Ahrens S, Zelenay S, Sancho D, Hanc P, Kjaer S, Feest C, Fletcher G, Durkin C, Postigo A, Skehel M, et al. F-actin is an evolutionarily conserved damage-associated molecular pattern recognized by DNGR-1, a receptor for dead cells. Immunity. 2012;36:635–645. - PubMed
-
- Collins BE, Paulson JC. Cell surface biology mediated by low affinity multivalent protein-glycan interactions. Current opinion in chemical biology. 2004;8:617–625. - PubMed
-
- Connor RJ, Kawaoka Y, Webster RG, Paulson JC. Receptor specificity in human, avian, and equine H2 and H3 influenza virus isolates. Virology. 1994;205:17–23. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
