An ancient ecospecies of Helicobacter pylori
- PMID: 39415013
- PMCID: PMC11541087
- DOI: 10.1038/s41586-024-07991-z
An ancient ecospecies of Helicobacter pylori
Abstract
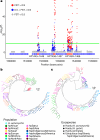
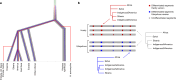
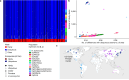
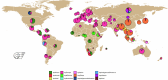
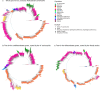
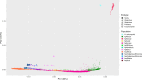
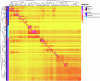
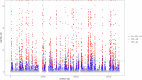
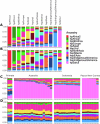
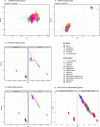
Helicobacter pylori disturbs the stomach lining during long-term colonization of its human host, with sequelae including ulcers and gastric cancer1,2. Numerous H. pylori virulence factors have been identified, showing extensive geographic variation1. Here we identify a 'Hardy' ecospecies of H. pylori that shares the ancestry of 'Ubiquitous' H. pylori from the same region in most of the genome but has nearly fixed single-nucleotide polymorphism differences in 100 genes, many of which encode outer membrane proteins and host interaction factors. Most Hardy strains have a second urease, which uses iron as a cofactor rather than nickel3, and two additional copies of the vacuolating cytotoxin VacA. Hardy strains currently have a limited distribution, including in Indigenous populations in Siberia and the Americas and in lineages that have jumped from humans to other mammals. Analysis of polymorphism data implies that Hardy and Ubiquitous coexisted in the stomachs of modern humans since before we left Africa and that both were dispersed around the world by our migrations. Our results also show that highly distinct adaptive strategies can arise and be maintained stably within bacterial populations, even in the presence of continuous genetic exchange between strains.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Suerbaum, S. & Michetti, P. Helicobacter pylori infection. N. Engl. J. Med.347, 1175–1186 (2002). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

