Geographics and bacterial networks differently shape the acquired and latent global sewage resistomes
- PMID: 41271719
- PMCID: PMC12639157
- DOI: 10.1038/s41467-025-66070-7
Geographics and bacterial networks differently shape the acquired and latent global sewage resistomes
Abstract
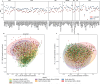
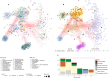
Antimicrobial resistance genes (ARGs) have rapidly emerged and spread globally, but the pathways driving their spread remain poorly understood. We analyzed 1240 sewage samples from 351 cities across 111 countries, comparing ARGs known to be mobilized with those identified through functional metagenomics (FG). FG ARGs showed stronger associations with bacterial taxa than the acquired ARGs. Network analyses further confirmed this and showed potential for source attribution of both known and novel ARGs. The FG resistome was more evenly dispersed globally, whereas the acquired resistome followed distinct geographical patterns. City-wise distance-decay analyses revealed that the FG ARGs showed significant decay within countries but not across regions or globally. In contrast, acquired ARGs showed decay at both national and regional scales. At the variant level, both ARG groups had significant national and regional distance-decay effects, but only FG ARGs at a global scale. Additionally, we observed stronger distance effects in Sub-Saharan Africa and East Asia compared to North America. Our findings suggest that differential selection and niche competition, rather than dispersal, shape the global resistome patterns. A limited number of bacterial taxa may act as reservoirs of latent FG ARGs, highlighting the need of targeted surveillance to mitigate future resistance threats.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
-
- One Health Global Leaders Group on Antimicrobial Resistance. GLG Report: Towards Specific Commitments and Action in the Response to Antimicrobial Resistance. https://www.amrleaders.org/about-us/what-we-do/glg-report (2024).
-
- D’Costa, V. M. et al. Antibiotic resistance is ancient. Nature477, 457–461 (2011). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

