Active residues and viral substrate cleavage sites of the protease of the birnavirus infectious pancreatic necrosis virus
- PMID: 10666235
- PMCID: PMC111686
- DOI: 10.1128/jvi.74.5.2057-2066.2000
Active residues and viral substrate cleavage sites of the protease of the birnavirus infectious pancreatic necrosis virus
Abstract
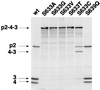
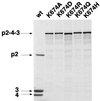
The polyprotein of infectious pancreatic necrosis virus (IPNV), a birnavirus, is processed by the viral protease VP4 (also named NS) to generate three polypeptides: pVP2, VP4, and VP3. Site-directed mutagenesis at 42 positions of the IPNV VP4 protein was performed to determine the active site and the important residues for the protease activity. Two residues (serine 633 and lysine 674) were critical for cleavage activity at both the pVP2-VP4 and the VP4-VP3 junctions. Wild-type activity at the pVP2-VP4 junction and a partial block (with an alteration of the cleavage specificity) at the VP4-VP3 junction were observed when replacement occurred at histidines 547 and 679. A similar observation was made when aspartic acid 693 was replaced by leucine, but wild-type activity and specificity were found when substituted by glutamine or asparagine. Sequence comparison between IPNV and two birnavirus (infectious bursal disease virus and Drosophila X virus) VP4s revealed that serine 633 and lysine 674 are conserved in these viruses, in contrast to histidines 547 and 679. The importance of serine 633 and lysine 674 is reminiscent of the protease active site of bacterial leader peptidases and their mitochondrial homologs and of the bacterial LexA-like proteases. Self-cleavage sites of IPNV VP4 were determined at the pVP2-VP4 and VP4-VP3 junctions by N-terminal sequencing and mutagenesis. Two alternative cleavage sites were also identified in the carboxyl domain of pVP2 by cumulative mutagenesis. The results suggest that VP4 cleaves the (Ser/Thr)-X-Ala / (Ser/Ala)-Gly motif, a target sequence with similarities to bacterial leader peptidases and herpesvirus protease cleavage sites.
Figures







References
-
- Allaire M, Chernaia M M, Malcolm B A, James M N G. Picornaviral 3C cysteine proteinases have a fold similar to chymotrypsin-like serine proteinases. Nature. 1994;369:72–76. - PubMed
-
- Allsop A E, Ashby M, Brooks G, Bruton G, Coulton S, Edwards P D, Elsmere S A, Hatton I K, Kaura A C, McLean S D, Pearson M J, Pearson N D, Perry C R, Smale T, Southgate R. Inhibition of protein export in bacteria: the signalling of a new role for β-lactams. In: Bentley P H, O'Hanlon P J, editors. Anti-infectives: recent advances in chemistry and structure-activity relationships—1997. Cambridge, England: The Royal Society of Chemistry; 1997. pp. 61–72.
-
- Azad A A, Jagadish M N, Brown M A, Hudson P J. Deletion mapping and expression in Escherichia coli of the large genomic segment of a birnavirus. Virology. 1987;161:145–152. - PubMed
-
- Babé L A, Craik C S. Viral proteases: evolution of diverse structural motifs to optimize function. Cell. 1997;91:427–430. - PubMed
-
- Barrett A J, Rawlings N D. Families and clans of serine peptidases. Arch Biochem Biophys. 1995;318:247–250. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

