Mdm12p, a component required for mitochondrial inheritance that is conserved between budding and fission yeast
- PMID: 9024686
- PMCID: PMC2134291
- DOI: 10.1083/jcb.136.3.545
Mdm12p, a component required for mitochondrial inheritance that is conserved between budding and fission yeast
Abstract
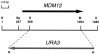
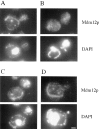
Saccharomyces cerevisiae cells lacking the MDM12 gene product display temperature-sensitive growth and possess abnormally large, round mitochondria that are defective for inheritance by daughter buds. Analysis of the wild-type MDM12 gene revealed its product to be a 31-kD polypeptide that is homologous to a protein of the fission yeast Schizosaccharomyces pombe. When expressed in S. cerevisiae, the S. pombe Mdm12p homolog conferred a dominant-negative phenotype of giant mitochondria and aberrant mitochondrial distribution, suggesting partial functional conservation of Mdm12p activity between budding and fission yeast. The S. cerevisiae Mdm12p was localized by indirect immunofluorescence microscopy and by subcellular fractionation and immunodetection to the mitochondrial outer membrane and displayed biochemical properties of an integral membrane protein. Mdm12p is the third mitochondrial outer membrane protein required for normal mitochondrial morphology and distribution to be identified in S. cerevisiae and the first such mitochondrial component that is conserved between two different species.
Figures







References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Attardi G, Schatz G. Biogenesis of mitochondria. Annu Rev Cell Biol. 1988;4:289–333. - PubMed
-
- Bereiter-Hahn J. Dimethylaminostyrylmethylpyridiniumiodine (DASPMI) as a fluorescent probe for mitochondria in situ. Biochim Biophys Acta. 1976;423:1–14. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

