Promiscuous translocations into immunoglobulin heavy chain switch regions in multiple myeloma
- PMID: 8943038
- PMCID: PMC19472
- DOI: 10.1073/pnas.93.24.13931
Promiscuous translocations into immunoglobulin heavy chain switch regions in multiple myeloma
Abstract
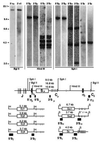
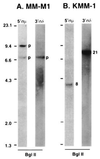
In multiple myeloma, karyotopic 14q32 translocations have been identified at a variable frequency (10-60% in different studies). In the majority of cases, the partner chromosome has not been identified (14q+), and in the remaining cases, a diverse array of chromosomal partners has been implicated, with 11q13 being the most common. We developed a comprehensive Southern blot assay to identify and distinguish different kinds of immunoglobulin heavy chain (IgH) switch recombination events. Illegitimate switch recombination fragments (defined as containing sequences from only one switch region) are potential markers of translocation events into IgH switch regions and were identified in 15 of 21 myeloma cell lines, including seven of eight karyotyped lines that have no detectable 14q32 translocation. From all nine lines or tumor samples analyzed further, cloned illegitimate switch recombination fragments were confirmed to be IgH switch translocation breakpoints. In three of these cases, the translocation breakpoint was shown to be present in the primary tumor. These translocation breakpoints involve six chromosomal loci: 4p16.3 (two lines and the one tumor); 6; 8q24.13; 11q13.3 (in three lines); 16q23.1; and 21q22.1. We suggest that translocations into the IgH locus (i) are frequent (karyotypic 14q32 translocations and/or illegitimate switch recombination fragments are present in primary tumor samples and in 19 of 21 lines that we have analyzed); (ii) occur mainly in switch regions; and (iii) involve a diverse but nonrandom array (i.e., frequently 11q13 or 4p16) of chromosomal partners. This appears to be the most frequent genetic abnormality in multiple myeloma.
Figures


References
-
- Malpas J S, Bergsagel D E, Kyle R. Multiple Myeloma: Biology and Management. Oxford: Oxford Univ. Press; 1995.
-
- Kyle R A. Mayo Clin Proc. 1993;68:26–36. - PubMed
-
- Lai J L, Zandecki M, Mary J Y, Bernardi F, Izydorczyk V, Flactif M, Morel P, Jouet J P, Bauters F, Facon T. Blood. 1995;85:2490–2497. - PubMed
-
- Sawyer J R, Waldron J A, Jagannath S, Barlogie B. Cancer Genet Cytogenet. 1995;82:41–49. - PubMed
-
- Taniwaki M, Nishida K, Takashima T, Nakagawa H, Fujii H, Tamaki T, Shimazaki C, Horiike S, Misawa S, Abe T, Kashima K. Blood. 1994;84:2283–2290. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

