Safety, tolerability, viral kinetics, and immune correlates of protection in healthy, seropositive UK adults inoculated with SARS-CoV-2: a single-centre, open-label, phase 1 controlled human infection study
- PMID: 38703782
- PMCID: PMC7616636
- DOI: 10.1016/S2666-5247(24)00025-9
Safety, tolerability, viral kinetics, and immune correlates of protection in healthy, seropositive UK adults inoculated with SARS-CoV-2: a single-centre, open-label, phase 1 controlled human infection study
Abstract
Background: A SARS-CoV-2 controlled human infection model (CHIM) has been successfully established in seronegative individuals using a dose of 1×101 50% tissue culture infectious dose (TCID50) pre-alpha SARS-CoV-2 virus. Given the increasing prevalence of seropositivity to SARS-CoV-2, a CHIM that could be used for vaccine development will need to induce infection in those with pre-existing immunity. Our aim was to find a dose of pre-alpha SARS-CoV-2 virus that induced infection in previously infected individuals.
Methods: Healthy, UK volunteers aged 18-30 years, with proven (quantitative RT-PCR or lateral flow antigen test) previous SARS-CoV-2 infection (with or without vaccination) were inoculated intranasally in a stepwise dose escalation CHIM with either 1×101, 1×102, 1×10³, 1×104, or 1×105 TCID50 SARS-CoV-2/human/GBR/484861/2020, the same virus used in the seronegative CHIM. Post-inoculation, volunteers were quarantined in functionally negative pressure rooms (Oxford, UK) for 14 days and until 12-hourly combined oropharyngeal-nasal swabs were negative for viable virus by focus-forming assay. Outpatient follow-up continued for 12 months post-enrolment, with additional visits for those who developed community-acquired SARS-CoV-2 infection. The primary objective was to identify a safe, well tolerated dose that induced infection (defined as two consecutive SARS-CoV-2 positive PCRs starting 24 h after inoculation) in 50% of seropositive volunteers. This study is registered with ClinicalTrials.gov (NCT04864548); enrolment and follow-up to 12 months post-enrolment are complete.
Findings: Recruitment commenced on May 6, 2021, with the last volunteer enrolled into the dose escalation cohort on Nov 24, 2022. 36 volunteers were enrolled, with four to eight volunteers inoculated in each dosing group from 1×101 to 1×105 TCID50 SARS-CoV-2. All volunteers have completed quarantine, with follow-up to 12 months complete. Despite dose escalation to 1×105 TCID50, we were unable to induce sustained infection in any volunteers. Five (14%) of 36 volunteers were considered to have transient infection, based on the kinetic of their PCR-positive swabs. Transiently infected volunteers had significantly lower baseline mucosal and systemic SARS-CoV-2-specific antibody titres and significantly lower peripheral IFNγ responses against a CD8+ T-cell SARS-CoV-2 peptide pool than uninfected volunteers. 14 (39%) of 36 volunteers subsequently developed breakthrough infection with the omicron variant after discharge from quarantine. Most adverse events reported by volunteers in quarantine were mild, with fatigue (16 [44%]) and stuffy nose (16 [44%]) being the most common. There were no serious adverse events.
Interpretation: Our study demonstrates potent protective immunity induced by homologous vaccination and homologous or heterologous previous SARS-CoV-2 infection. The community breakthrough infections seen with the omicron variant supports the use of newer variants to establish a model with sufficient rate of infection for use in vaccine and therapeutic development.
Funding: Wellcome Trust and Department for Health and Social Care.
Copyright © 2024 The Author(s). Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
Declaration of interests MC and SLa are supported by the Oak Foundation. TT, SLo, and SLa are supported by the US Food and Drug Administration Medical Countermeasures Initiative contract 75F40120C00085. VMF acknowledges support from the British Heart Foundation (CH/16/1/32013). AC is a full time paid employee of hVIVO. AJP is chair of the UK Department of Health and Social Care’s (DHSC) Joint Committee on Vaccination and Immunisation, was a member of WHO’s Strategic Advisory Group of Experts until 2022, receives consulting fees from Shionogi, and is in receipt of grants from the UK Medical Research Council, AstraZeneca, Gates Foundation, Wellcome Trust, National Institute for Health and Care Research (NIHR), Serum Institute of India, CEPI, and European Commission through the University of Oxford. AJP, SJ, HMo, SAH, RT, RL-R, and ICP contributed to intellectual property licensed by Oxford University Innovation to AstraZeneca. All other authors declare no competing interests.
Figures

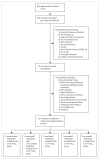

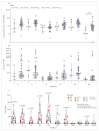
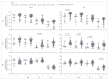
References
-
- Khoury DS, Cromer D, Reynaldi A, et al. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat Med. 2021;27:1205–11. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

