Regulatory interaction of PRL1 WD protein with Arabidopsis SNF1-like protein kinases
- PMID: 10220464
- PMCID: PMC21862
- DOI: 10.1073/pnas.96.9.5322
Regulatory interaction of PRL1 WD protein with Arabidopsis SNF1-like protein kinases
Abstract
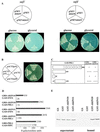
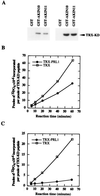
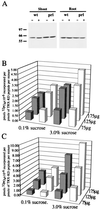
Mutation of the PRL1 gene, encoding a regulatory WD protein, results in glucose hypersensitivity and derepression of glucose-regulated genes in Arabidopsis. The yeast SNF1 protein kinase, a key regulator of glucose signaling, and Arabidopsis SNF1 homologs AKIN10 and AKIN11, which can complement the Deltasnf1 mutation, were found to interact with an N-terminal domain of the PRL1 protein in the two-hybrid system and in vitro. AKIN10 and AKIN11 suppress the yeast Deltasnf4 mutation and interact with the SNF4p-activating subunit of SNF1. PRL1 and SNF4 bind independently to adjacent C-terminal domains of AKIN10 and AKIN11, and these protein interactions are negatively regulated by glucose in yeast. AKIN10 and AKIN11, purified in fusion with glutathione S-transferase, undergo autophosphorylation and phosphorylate a peptide of sucrose phosphate synthase in vitro. The sucrose phosphate synthase-peptide kinase activity of AKIN complexes detected by immunoprecipitation is stimulated by sucrose in light-grown Arabidopsis plants. In comparison with wild type, the activation level of AKIN immunocomplexes is higher in the prl1 mutant, suggesting that PRL1 is a negative regulator of Arabidopsis SNF1 homologs. This conclusion is supported by the observation that PRL1 is an inhibitor of AKIN10 and AKIN11 in vitro.
Figures

Comment in
-
Another player joins the complex field of sugar-regulated gene expression in plants.Proc Natl Acad Sci U S A. 1999 Apr 27;96(9):4746-8. doi: 10.1073/pnas.96.9.4746. Proc Natl Acad Sci U S A. 1999. PMID: 10220362 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

