The N. gonorrhoeae type IV pilus stimulates mechanosensitive pathways and cytoprotection through a pilT-dependent mechanism
- PMID: 15769184
- PMCID: PMC1065265
- DOI: 10.1371/journal.pbio.0030100
The N. gonorrhoeae type IV pilus stimulates mechanosensitive pathways and cytoprotection through a pilT-dependent mechanism
Abstract
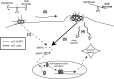
The Neisseria gonorrhoeae type IV pilus is a retractile appendage that can generate forces near 100 pN. We tested the hypothesis that type IV pilus retraction influences epithelial cell gene expression by exerting tension on the host membrane. Wild-type and retraction-defective bacteria altered the expression of an identical set of epithelial cell genes during attachment. Interestingly, pilus retraction, per se, did not regulate novel gene expression but, rather, enhanced the expression of a subset of the infection-regulated genes. This is accomplished through mitogen-activated protein kinase activation and at least one other undefined stress-activated pathway. These results can be reproduced by applying artificial force on the epithelial membrane, using a magnet and magnetic beads. Importantly, this retraction-mediated signaling increases the ability of the cell to withstand apoptotic signals triggered by infection. We conclude that pilus retraction stimulates mechanosensitive pathways that enhance the expression of stress-responsive genes and activate cytoprotective signaling. A model for the role of pilus retraction in influencing host cell survival is presented.
Figures

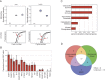
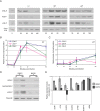
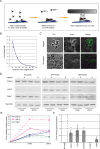
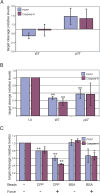
References
-
- Mattick JS. Type IV pili and twitching motility. Annu Rev Microbiol. 2002;56:289–314. - PubMed
-
- Wall D, Kaiser D. Type IV pili and cell motility. Mol Microbiol. 1999;32:1–10. - PubMed
-
- O'Toole GA, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol. 1998;30:295–304. - PubMed
-
- Bechet M, Blondeau R. Factors associated with the adherence and biofilm formation by Aeromonas caviae on glass surfaces. J Appl Microbiol. 2003;94:1072–1078. - PubMed
-
- Dubnau D. DNA uptake in bacteria. Annu Rev Microbiol. 1999;53:217–244. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

