Functional characterization of a multi-cancer risk locus on chr5p15.33 reveals regulation of TERT by ZNF148
- PMID: 28447668
- PMCID: PMC5414179
- DOI: 10.1038/ncomms15034
Functional characterization of a multi-cancer risk locus on chr5p15.33 reveals regulation of TERT by ZNF148
Erratum in
-
Publisher correction: Functional characterization of a multi-cancer risk locus on chr5p15.33 reveals regulation of TERT by ZNF148.Nat Commun. 2018 Mar 5;9:16159. doi: 10.1038/ncomms16159. Nat Commun. 2018. PMID: 29596408 Free PMC article.
Abstract
Genome wide association studies (GWAS) have mapped multiple independent cancer susceptibility loci to chr5p15.33. Here, we show that fine-mapping of pancreatic and testicular cancer GWAS within one of these loci (Region 2 in CLPTM1L) focuses the signal to nine highly correlated SNPs. Of these, rs36115365-C associated with increased pancreatic and testicular but decreased lung cancer and melanoma risk, and exhibited preferred protein-binding and enhanced regulatory activity. Transcriptional gene silencing of this regulatory element repressed TERT expression in an allele-specific manner. Proteomic analysis identifies allele-preferred binding of Zinc finger protein 148 (ZNF148) to rs36115365-C, further supported by binding of purified recombinant ZNF148. Knockdown of ZNF148 results in reduced TERT expression, telomerase activity and telomere length. Our results indicate that the association with chr5p15.33-Region 2 may be explained by rs36115365, a variant influencing TERT expression via ZNF148 in a manner consistent with elevated TERT in carriers of the C allele.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


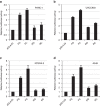

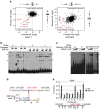
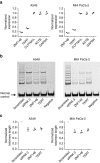
References
Publication types
MeSH terms
Substances
Grants and funding
- G1000143/MRC_/Medical Research Council/United Kingdom
- P30 CA023108/CA/NCI NIH HHS/United States
- 19167/CRUK_/Cancer Research UK/United Kingdom
- 14136/CRUK_/Cancer Research UK/United Kingdom
- G0401527/MRC_/Medical Research Council/United Kingdom
- R01 CA154823/CA/NCI NIH HHS/United States
- 10589/CRUK_/Cancer Research UK/United Kingdom
- UM1 CA167462/CA/NCI NIH HHS/United States
- MR/N003284/1/MRC_/Medical Research Council/United Kingdom
- P50 CA062924/CA/NCI NIH HHS/United States
- U01 CA164947/CA/NCI NIH HHS/United States
- UM1 CA182934/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

