Comparison of specific endophytic bacterial communities in different developmental stages of Passiflora incarnata using culture-dependent and culture-independent analysis
- PMID: 31454177
- PMCID: PMC6813437
- DOI: 10.1002/mbo3.896
Comparison of specific endophytic bacterial communities in different developmental stages of Passiflora incarnata using culture-dependent and culture-independent analysis
Abstract
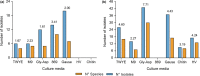
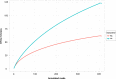
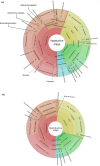
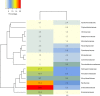
Plants and endophytic microorganisms have coevolved unique relationships over many generations. Plants show a specific physiological status in each developmental stage, which may determine the occurrence and dominance of specific endophytic populations with a predetermined ecological role. This study aimed to compare and determine the structure and composition of cultivable and uncultivable bacterial endophytic communities in vegetative and reproductive stages (RS) of Passiflora incarnata. To that end, the endophytic communities were assessed by plating and Illumina-based 16S rRNA gene amplicon sequencing. Two hundred and four cultivable bacterial strains were successfully isolated. From the plant's RS, the isolated strains were identified mainly as belonging to the genera Sphingomonas, Curtobacterium, and Methylobacterium, whereas Bacillus was the dominant genus isolated from the vegetative stage (VS). From a total of 133,399 sequences obtained from Illumina-based sequencing, a subset of 25,092 was classified in operational taxonomy units (OTUs). Four hundred and sixteen OTUs were obtained from the VS and 66 from the RS. In the VS, the most abundant families were Pseudoalteromonadaceae and Alicyclobacillaceae, while in the RS, Enterobacteriaceae and Bacillaceae were the most abundant families. The exclusive abundance of specific bacterial populations for each developmental stage suggests that plants may modulate bacterial endophytic community structure in response to different physiological statuses occurring at the different plant developmental stages.
Keywords: 16S rRNA gene sequencing; diversity; endophytic microbiome; microbial ecology; plant development.
© 2019 The Authors. MicrobiologyOpen published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that there is no conflict of interest.
Figures

References
-
- Andreolli, M. , Lampis, S. , Zapparoli, G. , Angelini, E. , & Vallini, G. (2016). Diversity of bacterial endophytes in 3 and 15 year‐old grapevines of Vitis vinifera cv. Corvina and their potential for plant growth promotion and phytopathogen control. Microbiological Research, 183, 42–52. - PubMed
-
- Andrews, S. (2012). FastQC: A quality control tool for high throughput sequence. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

